A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening
- PMID: 23386264
- PMCID: PMC3608766
- DOI: 10.1105/tpc.112.108118
A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening
Abstract
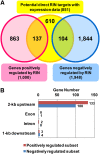
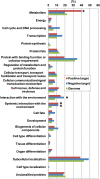
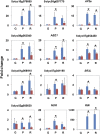
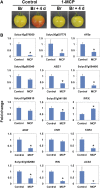
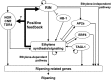
The fruit ripening developmental program is specific to plants bearing fleshy fruits and dramatically changes fruit characteristics, including color, aroma, and texture. The tomato (Solanum lycopersicum) MADS box transcription factor RIPENING INHIBITOR (RIN), one of the earliest acting ripening regulators, is required for both ethylene-dependent and -independent ripening regulatory pathways. Recent studies have identified two dozen direct RIN targets, but many more RIN targets remain to be identified. Here, we report the large-scale identification of direct RIN targets by chromatin immunoprecipitation coupled with DNA microarray analysis (ChIP-chip) targeting the predicted promoters of tomato genes. Our combined ChIP-chip and transcriptome analysis identified 241 direct RIN target genes that contain a RIN binding site and exhibit RIN-dependent positive or negative regulation during fruit ripening, suggesting that RIN has both activator and repressor roles. Examination of the predicted functions of RIN targets revealed that RIN participates in the regulation of lycopene accumulation, ethylene production, chlorophyll degradation, and many other physiological processes. Analysis of the effect of ethylene using 1-methylcyclopropene revealed that the positively regulated subset of RIN targets includes ethylene-sensitive and -insensitive transcription factors. Intriguingly, ethylene is involved in the upregulation of RIN expression during ripening. These results suggest that tomato fruit ripening is regulated by the interaction between RIN and ethylene signaling.
Figures


Similar articles
-
Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses.Planta. 2012 Jun;235(6):1107-22. doi: 10.1007/s00425-011-1561-2. Epub 2011 Dec 9. Planta. 2012. PMID: 22160566
-
The regulatory mechanism of fruit ripening revealed by analyses of direct targets of the tomato MADS-box transcription factor RIPENING INHIBITOR.Plant Signal Behav. 2013 Jun;8(6):e24357. doi: 10.4161/psb.24357. Epub 2013 Mar 21. Plant Signal Behav. 2013. PMID: 23518588 Free PMC article.
-
TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network.Plant J. 2009 Dec;60(6):1081-95. doi: 10.1111/j.1365-313X.2009.04064.x. Plant J. 2009. PMID: 19891701
-
Variations on a theme in fruit development: the PLE lineage of MADS-box genes in tomato (TAGL1) and other species.Planta. 2017 Aug;246(2):313-321. doi: 10.1007/s00425-017-2725-5. Epub 2017 Jun 28. Planta. 2017. PMID: 28660293 Review.
-
A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening.New Phytol. 2019 Mar;221(4):1724-1741. doi: 10.1111/nph.15545. Epub 2018 Nov 8. New Phytol. 2019. PMID: 30328615 Review.
Cited by
-
The NAC side of the fruit: tuning of fruit development and maturation.BMC Plant Biol. 2021 May 27;21(1):238. doi: 10.1186/s12870-021-03029-y. BMC Plant Biol. 2021. PMID: 34044765 Free PMC article. Review.
-
Polycomb-group protein SlMSI1 represses the expression of fruit-ripening genes to prolong shelf life in tomato.Sci Rep. 2016 Aug 25;6:31806. doi: 10.1038/srep31806. Sci Rep. 2016. PMID: 27558543 Free PMC article.
-
GWAS and transcriptome analysis reveal MADS26 involved in seed germination ability in maize.Theor Appl Genet. 2022 May;135(5):1717-1730. doi: 10.1007/s00122-022-04065-4. Epub 2022 Mar 5. Theor Appl Genet. 2022. PMID: 35247071
-
Red light-transmittance bagging promotes carotenoid accumulation of grapefruit during ripening.Commun Biol. 2022 Apr 4;5(1):303. doi: 10.1038/s42003-022-03270-7. Commun Biol. 2022. PMID: 35379890 Free PMC article.
-
Silencing of the Target of Rapamycin Complex Genes Stimulates Tomato Fruit Ripening.Mol Cells. 2022 Sep 30;45(9):660-672. doi: 10.14348/molcells.2022.2025. Epub 2022 Aug 22. Mol Cells. 2022. PMID: 35993163 Free PMC article.
References
-
- Chung M.Y., Vrebalov J., Alba R., Lee J., McQuinn R., Chung J.D., Klein P., Giovannoni J. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64: 936–947 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

