MSK1 and MSK2 inhibit lipopolysaccharide-induced prostaglandin production via an interleukin-10 feedback loop
- PMID: 23382072
- PMCID: PMC3624268
- DOI: 10.1128/MCB.01690-12
MSK1 and MSK2 inhibit lipopolysaccharide-induced prostaglandin production via an interleukin-10 feedback loop
Abstract
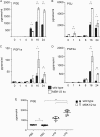
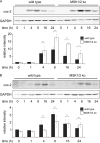
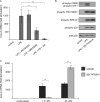
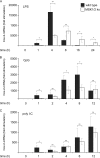
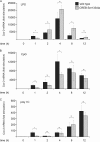
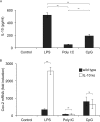
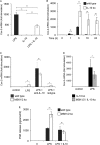
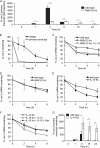
Prostaglandin production is catalyzed by cyclooxygenase 2 (cox-2). We demonstrate here that MSK1 and MSK2 (MSK1/2) can exert control on the induction of cox-2 mRNA by Toll-like receptor (TLR) agonists. In the initial phase of cox-2 induction, MSK1/2 knockout macrophages confirmed a role for MSK in the positive regulation of transcription. However, at later time points both lipopolysaccharide (LPS)-induced prostaglandin and cox-2 protein levels were increased in MSK1/2 knockout. Further analysis found that while MSKs promoted cox-2 mRNA transcription, following longer LPS stimulation MSKs also promoted degradation of cox-2 mRNA. This was found to be the result of an interleukin 10 (IL-10) feedback mechanism, with endogenously produced IL-10 promoting cox-2 degradation. The ability of IL-10 to do this was dependent on the mRNA binding protein TTP through a p38/MK2-mediated mechanism. As MSKs regulate IL-10 production in response to LPS, MSK1/2 knockout results in reduced IL-10 secretion and therefore reduced feedback from IL-10 on cox-2 mRNA stability. Following LPS stimulation, this increased mRNA stability correlated to an elevated induction of both of cox-2 protein and prostaglandin secretion in MSK1/2 knockout macrophages relative to that in wild-type cells. This was not restricted to isolated macrophages, as a similar effect of MSK1/2 knockout was seen on plasma prostaglandin E2 (PGE2) levels following intraperitoneal injection of LPS.
Figures

Similar articles
-
Beta Interferon Production Is Regulated by p38 Mitogen-Activated Protein Kinase in Macrophages via both MSK1/2- and Tristetraprolin-Dependent Pathways.Mol Cell Biol. 2016 Dec 19;37(1):e00454-16. doi: 10.1128/MCB.00454-16. Print 2017 Jan 1. Mol Cell Biol. 2016. PMID: 27795299 Free PMC article.
-
MSK1 regulates the transcription of IL-1ra in response to TLR activation in macrophages.Biochem J. 2010 Jan 15;425(3):595-602. doi: 10.1042/BJ20091062. Biochem J. 2010. PMID: 19922413
-
Dectin-1 regulates IL-10 production via a MSK1/2 and CREB dependent pathway and promotes the induction of regulatory macrophage markers.PLoS One. 2013;8(3):e60086. doi: 10.1371/journal.pone.0060086. Epub 2013 Mar 22. PLoS One. 2013. PMID: 23533666 Free PMC article.
-
Differential control of Toll-like receptor 4-induced interleukin-10 induction in macrophages and B cells reveals a role for p90 ribosomal S6 kinases.J Biol Chem. 2018 Feb 16;293(7):2302-2317. doi: 10.1074/jbc.M117.805424. Epub 2017 Dec 11. J Biol Chem. 2018. PMID: 29229781 Free PMC article.
-
The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling.Nat Immunol. 2008 Sep;9(9):1028-36. doi: 10.1038/ni.1644. Nat Immunol. 2008. PMID: 18690222
Cited by
-
Toll-like receptors in sepsis-associated cytokine storm and their endogenous negative regulators as future immunomodulatory targets.Int Immunopharmacol. 2020 Dec;89(Pt B):107087. doi: 10.1016/j.intimp.2020.107087. Epub 2020 Oct 12. Int Immunopharmacol. 2020. PMID: 33075714 Free PMC article. Review.
-
Beta Interferon Production Is Regulated by p38 Mitogen-Activated Protein Kinase in Macrophages via both MSK1/2- and Tristetraprolin-Dependent Pathways.Mol Cell Biol. 2016 Dec 19;37(1):e00454-16. doi: 10.1128/MCB.00454-16. Print 2017 Jan 1. Mol Cell Biol. 2016. PMID: 27795299 Free PMC article.
-
Staurosporine synergistically potentiates the deoxycholate-mediated induction of COX-2 expression.Physiol Rep. 2014 Aug 28;2(8):e12143. doi: 10.14814/phy2.12143. Print 2014 Aug 1. Physiol Rep. 2014. PMID: 25168879 Free PMC article.
-
Anti-inflammatory roles of p38α MAPK in macrophages are context dependent and require IL-10.J Leukoc Biol. 2017 Nov;102(5):1219-1227. doi: 10.1189/jlb.2AB0116-009RR. Epub 2017 Sep 6. J Leukoc Biol. 2017. PMID: 28877953 Free PMC article.
-
Phosphorylated Histone 3 at Serine 10 Identifies Activated Spinal Neurons and Contributes to the Development of Tissue Injury-Associated Pain.Sci Rep. 2017 Jan 25;7:41221. doi: 10.1038/srep41221. Sci Rep. 2017. PMID: 28120884 Free PMC article.
References
-
- Marnett LJ. 2009. The COXIB experience: a look in the rearview mirror. Annu. Rev. Pharmacol. Toxicol. 49: 265–290 - PubMed
-
- Smith WL, DeWitt DL, Garavito RM. 2000. Cyclooxygenases: structural, cellular, and molecular biology. Annu. Rev. Biochem. 69: 145–182 - PubMed
-
- Rajakariar R, Yaqoob MM, Gilroy DW. 2006. COX-2 in inflammation and resolution. Mol. Interv. 6: 199–207 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
