Structural basis for autoinhibition of the guanine nucleotide exchange factor FARP2
- PMID: 23375260
- PMCID: PMC3595398
- DOI: 10.1016/j.str.2013.01.001
Structural basis for autoinhibition of the guanine nucleotide exchange factor FARP2
Abstract
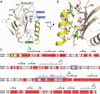
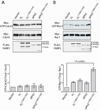
FARP2 is a Dbl-family guanine nucleotide exchange factor (GEF) that contains a 4.1, ezrin, radixin and moesin (FERM) domain, a Dbl-homology (DH) domain and two pleckstrin homology (PH) domains. FARP2 activates Rac1 or Cdc42 in response to upstream signals, thereby regulating processes such as neuronal axon guidance and bone homeostasis. How the GEF activity of FARP2 is regulated remained poorly understood. We have determined the crystal structures of the catalytic DH domain and the DH-PH-PH domains of FARP2. The structures reveal an auto-inhibited conformation in which the GEF substrate-binding site is blocked collectively by the last helix in the DH domain and the two PH domains. This conformation is stabilized by multiple interactions among the domains and two well-structured inter-domain linkers. Our cell-based activity assays confirm the suppression of the FARP2 GEF activity by these auto-inhibitory elements.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures




Similar articles
-
A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.EMBO J. 2002 Mar 15;21(6):1315-26. doi: 10.1093/emboj/21.6.1315. EMBO J. 2002. PMID: 11889037 Free PMC article.
-
The Tiam1 guanine nucleotide exchange factor is auto-inhibited by its pleckstrin homology coiled-coil extension domain.J Biol Chem. 2017 Oct 27;292(43):17777-17793. doi: 10.1074/jbc.M117.799114. Epub 2017 Sep 7. J Biol Chem. 2017. PMID: 28882897 Free PMC article.
-
Identification of Rho GTPase-dependent sites in the Dbl homology domain of oncogenic Dbl that are required for transformation.J Biol Chem. 2000 Aug 25;275(34):25993-6001. doi: 10.1074/jbc.M003780200. J Biol Chem. 2000. PMID: 10854437
-
New Mechanisms Underlying Oncogenesis in Dbl Family Rho Guanine Nucleotide Exchange Factors.Mol Pharmacol. 2024 Aug 16;106(3):117-128. doi: 10.1124/molpharm.124.000904. Mol Pharmacol. 2024. PMID: 38902036 Review.
-
The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling.Cell Signal. 2014 Mar;26(3):483-91. doi: 10.1016/j.cellsig.2013.11.034. Epub 2013 Dec 2. Cell Signal. 2014. PMID: 24308970 Review.
Cited by
-
FARP1 boosts CDC42 activity from integrin αvβ5 signaling and correlates with poor prognosis of advanced gastric cancer.Oncogenesis. 2020 Feb 6;9(2):13. doi: 10.1038/s41389-020-0190-7. Oncogenesis. 2020. PMID: 32029704 Free PMC article.
-
Regulation of DENND3, the exchange factor for the small GTPase Rab12 through an intramolecular interaction.J Biol Chem. 2017 Apr 28;292(17):7274-7282. doi: 10.1074/jbc.M116.772434. Epub 2017 Mar 1. J Biol Chem. 2017. PMID: 28249939 Free PMC article.
-
Integration of Chemoinformatics and Multi-Omics Analysis Defines ECT2 as a Potential Target for Cancer Drug Therapy.Biology (Basel). 2023 Apr 18;12(4):613. doi: 10.3390/biology12040613. Biology (Basel). 2023. PMID: 37106813 Free PMC article.
-
Membrane and Protein Interactions of the Pleckstrin Homology Domain Superfamily.Membranes (Basel). 2015 Oct 23;5(4):646-63. doi: 10.3390/membranes5040646. Membranes (Basel). 2015. PMID: 26512702 Free PMC article.
-
A CDC42-centered signaling unit is a dominant positive regulator of endothelial integrity.Sci Rep. 2017 Aug 31;7(1):10132. doi: 10.1038/s41598-017-10392-0. Sci Rep. 2017. PMID: 28860633 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta crystallographica. 2002;58:1948–1954. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

