Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs
- PMID: 23341450
- PMCID: PMC3597817
- DOI: 10.1074/jbc.M112.441907
Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs
Abstract
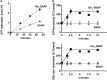
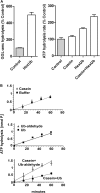
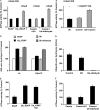
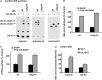
Degradation of ubiquitinated proteins by 26 S proteasomes requires ATP hydrolysis, but it is unclear how the proteasomal ATPases are regulated and how proteolysis, substrate deubiquitination, degradation, and ATP hydrolysis are coordinated. Polyubiquitinated proteins were shown to stimulate ATP hydrolysis by purified proteasomes, but only if the proteins contain a loosely folded domain. If they were not ubiquitinated, such proteins did not increase ATPase activity. However, they did so upon addition of ubiquitin aldehyde, which mimics the ubiquitin chain and binds to 26 S-associated deubiquitinating enzymes (DUBs): in yeast to Ubp6, which is essential for the ATPase activation, and in mammalian 26 S to the Ubp6 homolog, Usp14, and Uch37. Occupancy of either DUB by a ubiquitin conjugate leads to ATPase stimulation, thereby coupling deubiquitination and ATP hydrolysis. Thus, ubiquitinated loosely folded proteins, after becoming bound to the 26 S, interact with Ubp6/Usp14 or Uch37 to activate ATP hydrolysis and enhance their own destruction.
Figures

Similar articles
-
The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis.J Biol Chem. 2017 Jun 9;292(23):9830-9839. doi: 10.1074/jbc.M116.763128. Epub 2017 Apr 17. J Biol Chem. 2017. PMID: 28416611 Free PMC article.
-
UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells.Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11642-E11650. doi: 10.1073/pnas.1808731115. Epub 2018 Nov 28. Proc Natl Acad Sci U S A. 2018. PMID: 30487212 Free PMC article.
-
Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening.Mol Cell. 2009 Dec 11;36(5):794-804. doi: 10.1016/j.molcel.2009.11.015. Mol Cell. 2009. PMID: 20005843 Free PMC article.
-
Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes.Mol Cell Proteomics. 2011 May;10(5):R110.003871. doi: 10.1074/mcp.R110.003871. Epub 2010 Sep 7. Mol Cell Proteomics. 2011. PMID: 20823120 Free PMC article. Review.
-
Meddling with Fate: The Proteasomal Deubiquitinating Enzymes.J Mol Biol. 2017 Nov 10;429(22):3525-3545. doi: 10.1016/j.jmb.2017.09.015. Epub 2017 Oct 5. J Mol Biol. 2017. PMID: 28988953 Free PMC article. Review.
Cited by
-
Structural basis for dynamic regulation of the human 26S proteasome.Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):12991-12996. doi: 10.1073/pnas.1614614113. Epub 2016 Oct 21. Proc Natl Acad Sci U S A. 2016. PMID: 27791164 Free PMC article.
-
Specific lid-base contacts in the 26s proteasome control the conformational switching required for substrate degradation.Elife. 2019 Nov 28;8:e49806. doi: 10.7554/eLife.49806. Elife. 2019. PMID: 31778111 Free PMC article.
-
Measurement of the Multiple Activities of 26S Proteasomes.Methods Mol Biol. 2018;1844:289-308. doi: 10.1007/978-1-4939-8706-1_19. Methods Mol Biol. 2018. PMID: 30242717 Free PMC article.
-
Disassembly of Lys11 and mixed linkage polyubiquitin conjugates provides insights into function of proteasomal deubiquitinases Rpn11 and Ubp6.J Biol Chem. 2015 Feb 20;290(8):4688-4704. doi: 10.1074/jbc.M114.568295. Epub 2014 Nov 11. J Biol Chem. 2015. PMID: 25389291 Free PMC article.
-
Reversible phosphorylation of the 26S proteasome.Protein Cell. 2017 Apr;8(4):255-272. doi: 10.1007/s13238-017-0382-x. Epub 2017 Mar 3. Protein Cell. 2017. PMID: 28258412 Free PMC article. Review.
References
-
- Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 - PubMed
-
- Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. (2004) Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279, 26817–26822 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

