The NuRD architecture
- PMID: 23340908
- PMCID: PMC3652912
- DOI: 10.1007/s00018-012-1256-2
The NuRD architecture
Abstract
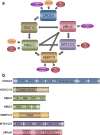
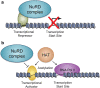
The nucleosome remodeling and deacetylase (NuRD) complex regulates chromatin organization, gene transcription, genomic stability and developmental signaling. NuRD has a unique dual enzymatic activity, containing an ATPase and a histone deacetylase among its six core subunits. Recent studies indicate that NuRD composition and the interplay between subunits may dictate the diverse functions of the complex. In this review, we examine the structures and biological roles of the NuRD subunits and discuss new avenues of research to advance our understanding of the NuRD-mediated signaling network.
Figures


Similar articles
-
CHD4 Is a Peripheral Component of the Nucleosome Remodeling and Deacetylase Complex.J Biol Chem. 2016 Jul 22;291(30):15853-66. doi: 10.1074/jbc.M115.707018. Epub 2016 May 27. J Biol Chem. 2016. PMID: 27235397 Free PMC article.
-
The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer.Epigenetics. 2009 Nov 16;4(8):532-6. doi: 10.4161/epi.4.8.10108. Epub 2009 Nov 16. Epigenetics. 2009. PMID: 19923891 Review.
-
The Nucleosome Remodeling and Deacetylase Complex NuRD Is Built from Preformed Catalytically Active Sub-modules.J Mol Biol. 2016 Jul 17;428(14):2931-42. doi: 10.1016/j.jmb.2016.04.025. Epub 2016 Apr 23. J Mol Biol. 2016. PMID: 27117189 Free PMC article.
-
Acetylation of histone deacetylase 1 regulates NuRD corepressor complex activity.J Biol Chem. 2012 Nov 23;287(48):40279-91. doi: 10.1074/jbc.M112.349704. Epub 2012 Sep 25. J Biol Chem. 2012. PMID: 23014989 Free PMC article.
-
The NuRD Complex in Neurodevelopment and Disease: A Case of Sliding Doors.Cells. 2023 Apr 18;12(8):1179. doi: 10.3390/cells12081179. Cells. 2023. PMID: 37190088 Free PMC article. Review.
Cited by
-
Transcription Factor hDREF Is a Novel SUMO E3 Ligase of Mi2α.J Biol Chem. 2016 May 27;291(22):11619-34. doi: 10.1074/jbc.M115.713370. Epub 2016 Apr 11. J Biol Chem. 2016. PMID: 27068747 Free PMC article.
-
Characterization of nucleosome sediments for protein interaction studies by solid-state NMR spectroscopy.Magn Reson (Gott). 2021;2(1):187-202. doi: 10.5194/mr-2-187-2021. Epub 2021 Apr 21. Magn Reson (Gott). 2021. PMID: 35647606 Free PMC article.
-
Functions and Interactions of Mammalian KDM5 Demethylases.Front Genet. 2022 Jul 11;13:906662. doi: 10.3389/fgene.2022.906662. eCollection 2022. Front Genet. 2022. PMID: 35899196 Free PMC article. Review.
-
The Tudor domain protein Spindlin1 is involved in intrinsic antiviral defense against incoming hepatitis B Virus and herpes simplex virus type 1.PLoS Pathog. 2014 Sep 11;10(9):e1004343. doi: 10.1371/journal.ppat.1004343. eCollection 2014 Sep. PLoS Pathog. 2014. PMID: 25211330 Free PMC article.
-
Regulation of Sufu activity by p66β and Mycbp provides new insight into vertebrate Hedgehog signaling.Genes Dev. 2014 Nov 15;28(22):2547-63. doi: 10.1101/gad.249425.114. Genes Dev. 2014. PMID: 25403183 Free PMC article.
References
-
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. - PubMed
-
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. - PubMed
-
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. - PubMed
-
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. - PubMed
-
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

