The structure of human microplasmin in complex with textilinin-1, an aprotinin-like inhibitor from the Australian brown snake
- PMID: 23335990
- PMCID: PMC3545990
- DOI: 10.1371/journal.pone.0054104
The structure of human microplasmin in complex with textilinin-1, an aprotinin-like inhibitor from the Australian brown snake
Abstract
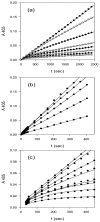
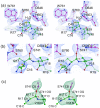
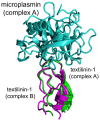
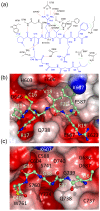
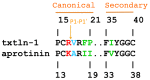
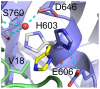
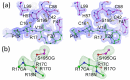
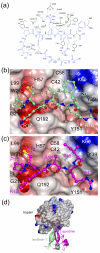
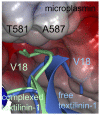
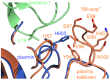
Textilinin-1 is a Kunitz-type serine protease inhibitor from Australian brown snake venom. Its ability to potently and specifically inhibit human plasmin (K(i) = 0.44 nM) makes it a potential therapeutic drug as a systemic anti-bleeding agent. The crystal structures of the human microplasmin-textilinin-1 and the trypsin-textilinin-1 complexes have been determined to 2.78 Å and 1.64 Å resolution respectively, and show that textilinin-1 binds to trypsin in a canonical mode but to microplasmin in an atypical mode with the catalytic histidine of microplasmin rotated out of the active site. The space vacated by the histidine side-chain in this complex is partially occupied by a water molecule. In the structure of microplasminogen the χ(1) dihedral angle of the side-chain of the catalytic histidine is rotated by 67° from its "active" position in the catalytic triad, as exemplified by its location when microplasmin is bound to streptokinase. However, when textilinin-1 binds to microplasmin the χ(1) dihedral angle of this amino acid residue changes by -157° (i.e. in the opposite rotation direction compared to microplasminogen). The unusual mode of interaction between textilinin-1 and plasmin explains textilinin-1's selectivity for human plasmin over plasma kallikrein. This difference can be exploited in future drug design efforts.
Conflict of interest statement
Figures

Similar articles
-
Crystal structure of textilinin-1, a Kunitz-type serine protease inhibitor from the venom of the Australian common brown snake (Pseudonaja textilis).FEBS J. 2009 Jun;276(11):3163-75. doi: 10.1111/j.1742-4658.2009.07034.x. Epub 2009 Apr 28. FEBS J. 2009. PMID: 19490116
-
Comparison of textilinin-1 with aprotinin as serine protease inhibitors and as antifibrinolytic agents.Pathophysiol Haemost Thromb. 2005;34(4-5):188-93. doi: 10.1159/000092421. Pathophysiol Haemost Thromb. 2005. PMID: 16707925
-
Crystallization and preliminary X-ray analysis of a Kunitz-type inhibitor, textilinin-1 from Pseudonaja textilis textilis.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006 Jul 1;62(Pt 7):642-5. doi: 10.1107/S1744309106019099. Epub 2006 Jun 10. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006. PMID: 16820682 Free PMC article.
-
Drug development from Australian elapid snake venoms and the Venomics pipeline of candidates for haemostasis: Textilinin-1 (Q8008), Haempatch™ (Q8009) and CoVase™ (V0801).Toxicon. 2012 Mar 15;59(4):456-63. doi: 10.1016/j.toxicon.2010.12.010. Epub 2010 Dec 22. Toxicon. 2012. PMID: 21184772 Review.
-
Bovine pancreatic trypsin inhibitor as a probe of large conductance Ca(2+)-activated K+ channels at an internal site of interaction.Biochem Pharmacol. 1992 Jan 9;43(1):21-8. doi: 10.1016/0006-2952(92)90656-4. Biochem Pharmacol. 1992. PMID: 1370897 Review.
Cited by
-
Identification, Characterization and Synthesis of Walterospermin, a Sperm Motility Activator from the Egyptian Black Snake Walterinnesia aegyptia Venom.Int J Mol Sci. 2020 Oct 21;21(20):7786. doi: 10.3390/ijms21207786. Int J Mol Sci. 2020. PMID: 33096770 Free PMC article.
-
A Review of Rattlesnake Venoms.Toxins (Basel). 2023 Dec 19;16(1):2. doi: 10.3390/toxins16010002. Toxins (Basel). 2023. PMID: 38276526 Free PMC article. Review.
-
Recent advances on plasmin inhibitors for the treatment of fibrinolysis-related disorders.Med Res Rev. 2014 Nov;34(6):1168-216. doi: 10.1002/med.21315. Epub 2014 Mar 21. Med Res Rev. 2014. PMID: 24659483 Free PMC article. Review.
-
Plant-Derived Compounds and Extracts as Modulators of Plasmin Activity-A Review.Molecules. 2023 Feb 9;28(4):1677. doi: 10.3390/molecules28041677. Molecules. 2023. PMID: 36838662 Free PMC article. Review.
-
Potent, Selective, Allosteric Inhibition of Human Plasmin by Sulfated Non-Saccharide Glycosaminoglycan Mimetics.J Med Chem. 2017 Jan 26;60(2):641-657. doi: 10.1021/acs.jmedchem.6b01474. Epub 2017 Jan 5. J Med Chem. 2017. PMID: 27976897 Free PMC article.
References
-
- Fergusson DA, Hebert PC, Mazer CD, Fremes S, MacAdams C, et al. (2008) A comparison of aprotinin and lysine analogues in high-risk cardiac surgery. N Engl J Med 358: 2319–2331. - PubMed
-
- Mangano DT, Tudor IC, Dietzel C (2006) The risk associated with aprotinin in cardiac surgery. N Engl J Med 354: 353–365. - PubMed
-
- Beattie WS, Karkouti K (2011) The post-BART anti-fibrinolytic dilemma? J Cardiothorac Vasc Anesth 25: 3–5. - PubMed
-
- Lewis RJ, Garcia ML (2003) Therapeutic potential of venom peptides. Nat Rev Drug Discov 2: 790–802. - PubMed
-
- Fry BG, Scheib H, van der Weerd L, Young B, McNaughtan J, et al. (2008) Evolution of an arsenal: structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol Cell Proteomics 7: 215–246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

