Posttranslational modification and quality control
- PMID: 23329792
- PMCID: PMC3566557
- DOI: 10.1161/CIRCRESAHA.112.268706
Posttranslational modification and quality control
Abstract
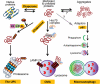
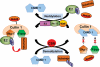
Protein quality control functions to minimize the level and toxicity of misfolded proteins in the cell. Protein quality control is performed by intricate collaboration among chaperones and target protein degradation. The latter is performed primarily by the ubiquitin-proteasome system and perhaps autophagy. Terminally misfolded proteins that are not timely removed tend to form aggregates. Their clearance requires macroautophagy. Macroautophagy serves in intracellular quality control also by selectively segregating defective organelles (eg, mitochondria) and targeting them for degradation by the lysosome. Inadequate protein quality control is observed in a large subset of failing human hearts with a variety of causes, and its pathogenic role has been experimentally demonstrated. Multiple posttranslational modifications can occur to substrate proteins and protein quality control machineries, promoting or hindering the removal of the misfolded proteins. This article highlights recent advances in posttranslational modification-mediated regulation of intracellular quality control mechanisms and its known involvement in cardiac pathology.
Figures
Similar articles
-
A protein quality control pathway at the mitochondrial outer membrane.Elife. 2020 Mar 2;9:e51065. doi: 10.7554/eLife.51065. Elife. 2020. PMID: 32118579 Free PMC article.
-
Ubiquitin receptors and protein quality control.J Mol Cell Cardiol. 2013 Feb;55:73-84. doi: 10.1016/j.yjmcc.2012.09.012. Epub 2012 Oct 6. J Mol Cell Cardiol. 2013. PMID: 23046644 Free PMC article. Review.
-
Autophagy: links with the proteasome.Curr Opin Cell Biol. 2010 Apr;22(2):192-8. doi: 10.1016/j.ceb.2009.11.002. Epub 2009 Dec 2. Curr Opin Cell Biol. 2010. PMID: 19962293
-
Targeting protein quality control pathways in breast cancer.BMC Biol. 2017 Nov 16;15(1):109. doi: 10.1186/s12915-017-0449-4. BMC Biol. 2017. PMID: 29145850 Free PMC article. Review.
-
[Molecular mechanisms and functions of autophagy and the ubiquitin-proteasome pathway].Yi Chuan. 2012 Jan;34(1):5-18. doi: 10.3724/sp.j.1005.2012.00005. Yi Chuan. 2012. PMID: 22306868 Review. Chinese.
Cited by
-
Highly Dynamic Changes in the Activity and Regulation of Macroautophagy in Hearts Subjected to Increased Proteotoxic Stress.Front Physiol. 2019 Jun 26;10:758. doi: 10.3389/fphys.2019.00758. eCollection 2019. Front Physiol. 2019. PMID: 31297061 Free PMC article.
-
Systemic inhibition of neddylation by 3-day MLN4924 treatment regime does not impair autophagic flux in mouse hearts and brains.Am J Cardiovasc Dis. 2017 Dec 20;7(6):134-150. eCollection 2017. Am J Cardiovasc Dis. 2017. PMID: 29348974 Free PMC article.
-
Recent Development of Genetic Code Expansion for Posttranslational Modification Studies.Molecules. 2018 Jul 8;23(7):1662. doi: 10.3390/molecules23071662. Molecules. 2018. PMID: 29986538 Free PMC article. Review.
-
Editorial: Post-translational Modifications and Compartmentalized Protein Quality Control in Cardiac Muscle and Disease.Front Physiol. 2021 Aug 30;12:745887. doi: 10.3389/fphys.2021.745887. eCollection 2021. Front Physiol. 2021. PMID: 34526915 Free PMC article. No abstract available.
-
Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins.Circulation. 2013 Jul 23;128(4):365-76. doi: 10.1161/CIRCULATIONAHA.113.001971. Epub 2013 Jun 14. Circulation. 2013. PMID: 23770744 Free PMC article.
References
-
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

