Timeless-dependent DNA replication-coupled recombination promotes Kaposi's Sarcoma-associated herpesvirus episome maintenance and terminal repeat stability
- PMID: 23325691
- PMCID: PMC3624199
- DOI: 10.1128/JVI.02211-12
Timeless-dependent DNA replication-coupled recombination promotes Kaposi's Sarcoma-associated herpesvirus episome maintenance and terminal repeat stability
Abstract
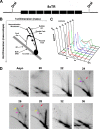
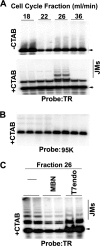
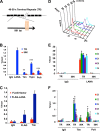
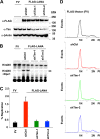
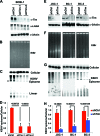
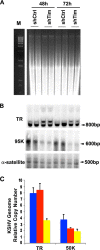
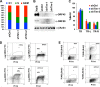
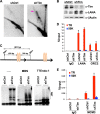
Kaposi's Sarcoma-associated herpesvirus (KSHV) is maintained as a stable episome in latently infected pleural effusion lymphoma (PEL) cells. Episome maintenance is conferred by the binding of the KSHV-encoded LANA protein to the viral terminal repeats (TR). Here, we show that DNA replication in the KSHV TR is coupled with DNA recombination and mediated in part through the cellular replication fork protection factors Timeless (Tim) and Tipin. We show by two-dimensional (2D) agarose gel electrophoresis that replication forks naturally stall and form recombination-like structures at the TR during an unperturbed cell cycle. Chromatin immunoprecipitation (ChIP) assays revealed that Tim and Tipin are selectively enriched at the KSHV TR during S phase and in a LANA-dependent manner. Tim depletion inhibited LANA-dependent TR DNA replication and caused the loss of KSHV episomes from latently infected PEL cells. Tim depletion resulted in the aberrant accumulation of recombination structures and arrested MCM helicase at TR. Tim depletion did not induce the KSHV lytic cycle or apoptotic cell death. We propose that KSHV episome maintenance requires Tim-assisted replication fork protection at the viral terminal repeats and that Tim-dependent recombination-like structures form at TR to promote DNA repeat stability and viral genome maintenance.
Figures
Similar articles
-
Identification of Kaposi's sarcoma-associated herpesvirus LANA regions important for episome segregation, replication, and persistence.J Virol. 2013 Nov;87(22):12270-83. doi: 10.1128/JVI.01243-13. Epub 2013 Sep 4. J Virol. 2013. PMID: 24006437 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus LANA-Adjacent Regions with Distinct Functions in Episome Segregation or Maintenance.J Virol. 2019 Mar 5;93(6):e02158-18. doi: 10.1128/JVI.02158-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626680 Free PMC article.
-
Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats.J Virol. 2006 Mar;80(5):2243-56. doi: 10.1128/JVI.80.5.2243-2256.2006. J Virol. 2006. PMID: 16474132 Free PMC article.
-
The latency-associated nuclear antigen, a multifunctional protein central to Kaposi's sarcoma-associated herpesvirus latency.Future Microbiol. 2011 Dec;6(12):1399-413. doi: 10.2217/fmb.11.137. Future Microbiol. 2011. PMID: 22122438 Free PMC article. Review.
-
KSHV Genome Replication and Maintenance.Front Microbiol. 2016 Feb 1;7:54. doi: 10.3389/fmicb.2016.00054. eCollection 2016. Front Microbiol. 2016. PMID: 26870016 Free PMC article. Review.
Cited by
-
Productive replication of human papillomavirus 31 requires DNA repair factor Nbs1.J Virol. 2014 Aug;88(15):8528-44. doi: 10.1128/JVI.00517-14. Epub 2014 May 21. J Virol. 2014. PMID: 24850735 Free PMC article.
-
ERK-mediated TIMELESS expression suppresses G2/M arrest in colon cancer cells.PLoS One. 2019 Jan 10;14(1):e0209224. doi: 10.1371/journal.pone.0209224. eCollection 2019. PLoS One. 2019. PMID: 30629587 Free PMC article.
-
Modulation of the DNA damage response during the life cycle of human papillomaviruses.Virus Res. 2017 Mar 2;231:41-49. doi: 10.1016/j.virusres.2016.11.006. Epub 2016 Nov 9. Virus Res. 2017. PMID: 27836727 Free PMC article. Review.
-
The Modulation of Apoptotic Pathways by Gammaherpesviruses.Front Microbiol. 2016 Apr 27;7:585. doi: 10.3389/fmicb.2016.00585. eCollection 2016. Front Microbiol. 2016. PMID: 27199919 Free PMC article. Review.
-
Kaposi's sarcoma-associated herpesvirus LANA recruits the DNA polymerase clamp loader to mediate efficient replication and virus persistence.Proc Natl Acad Sci U S A. 2014 Aug 12;111(32):11816-21. doi: 10.1073/pnas.1404219111. Epub 2014 Jul 28. Proc Natl Acad Sci U S A. 2014. PMID: 25071216 Free PMC article.
References
-
- Parkin DM. 2006. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 118:3030–3044 - PubMed
-
- Butel JS, Fan H. 2012. The diversity of human cancer viruses. Curr. Opin. Virol. 2:449–452 - PubMed
-
- Martin D, Gutkind JS. 2008. Human tumor-associated viruses and new insights into the molecular mechanisms of cancer. Oncogene 27(Suppl. 2):S31–S42 - PubMed
-
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

