A conserved mammalian protein interaction network
- PMID: 23320073
- PMCID: PMC3539715
- DOI: 10.1371/journal.pone.0052581
A conserved mammalian protein interaction network
Abstract
Physical interactions between proteins mediate a variety of biological functions, including signal transduction, physical structuring of the cell and regulation. While extensive catalogs of such interactions are known from model organisms, their evolutionary histories are difficult to study given the lack of interaction data from phylogenetic outgroups. Using phylogenomic approaches, we infer a upper bound on the time of origin for a large set of human protein-protein interactions, showing that most such interactions appear relatively ancient, dating no later than the radiation of placental mammals. By analyzing paired alignments of orthologous and putatively interacting protein-coding genes from eight mammals, we find evidence for weak but significant co-evolution, as measured by relative selective constraint, between pairs of genes with interacting proteins. However, we find no strong evidence for shared instances of directional selection within an interacting pair. Finally, we use a network approach to show that the distribution of selective constraint across the protein interaction network is non-random, with a clear tendency for interacting proteins to share similar selective constraints. Collectively, the results suggest that, on the whole, protein interactions in mammals are under selective constraint, presumably due to their functional roles.
Conflict of interest statement
Figures
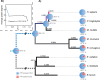
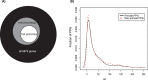
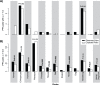
Similar articles
-
Human protein-RNA interaction network is highly stable across mammals.BMC Genomics. 2019 Dec 30;20(Suppl 12):1004. doi: 10.1186/s12864-019-6330-9. BMC Genomics. 2019. PMID: 31888461 Free PMC article.
-
Detection of nonneutral substitution rates on mammalian phylogenies.Genome Res. 2010 Jan;20(1):110-21. doi: 10.1101/gr.097857.109. Epub 2009 Oct 26. Genome Res. 2010. PMID: 19858363 Free PMC article.
-
Identification of constrained sequence elements across 239 primate genomes.Nature. 2024 Jan;625(7996):735-742. doi: 10.1038/s41586-023-06798-8. Epub 2023 Nov 29. Nature. 2024. PMID: 38030727 Free PMC article.
-
Mammalian evolution and biomedicine: new views from phylogeny.Biol Rev Camb Philos Soc. 2007 Aug;82(3):375-92. doi: 10.1111/j.1469-185X.2007.00016.x. Biol Rev Camb Philos Soc. 2007. PMID: 17624960 Review.
-
Support for interordinal eutherian relationships with an emphasis on primates and their archontan relatives.Mol Phylogenet Evol. 1996 Feb;5(1):78-88. doi: 10.1006/mpev.1996.0007. Mol Phylogenet Evol. 1996. PMID: 8673299 Review.
Cited by
-
Functional genomics of human brain development and implications for autism spectrum disorders.Transl Psychiatry. 2015 Oct 27;5(10):e665. doi: 10.1038/tp.2015.153. Transl Psychiatry. 2015. PMID: 26506051 Free PMC article. Review.
-
Intron Length Coevolution across Mammalian Genomes.Mol Biol Evol. 2016 Oct;33(10):2682-91. doi: 10.1093/molbev/msw151. Epub 2016 Aug 22. Mol Biol Evol. 2016. PMID: 27550903 Free PMC article.
-
Measuring protein interactions using Förster resonance energy transfer and fluorescence lifetime imaging microscopy.Methods. 2014 Mar 15;66(2):200-7. doi: 10.1016/j.ymeth.2013.06.017. Epub 2013 Jun 24. Methods. 2014. PMID: 23806643 Free PMC article.
-
Evolution of protein-protein interaction networks in yeast.PLoS One. 2017 Mar 1;12(3):e0171920. doi: 10.1371/journal.pone.0171920. eCollection 2017. PLoS One. 2017. PMID: 28248977 Free PMC article.
-
Recent positive selection has acted on genes encoding proteins with more interactions within the whole human interactome.Genome Biol Evol. 2015 Apr 2;7(4):1141-54. doi: 10.1093/gbe/evv055. Genome Biol Evol. 2015. PMID: 25840415 Free PMC article.
References
-
- Zhu X, Gerstein M, Snyder M (2007) Getting connected: analysis and principles of biological networks. Genes Dev 21: 1010–1024. - PubMed
-
- Teichmann SA, Babu MM (2004) Gene regulatory network growth by duplication. Nat Genet 36: 492–496. - PubMed
-
- Wagner A (2001) The yeast protein interaction network evolves rapidly and contains few redundant duplicate genes. Mol Biol Evol 18: 1283–1292. - PubMed
-
- Li L, Huang Y, Xia X, Sun Z (2006) Preferential duplication in the sparse part of yeast protein interaction network. Mol Biol Evol 23: 2467–2473. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

