Dual-site interactions of p53 protein transactivation domain with anti-apoptotic Bcl-2 family proteins reveal a highly convergent mechanism of divergent p53 pathways
- PMID: 23316052
- PMCID: PMC3591646
- DOI: 10.1074/jbc.M112.400754
Dual-site interactions of p53 protein transactivation domain with anti-apoptotic Bcl-2 family proteins reveal a highly convergent mechanism of divergent p53 pathways
Abstract
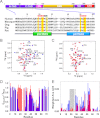
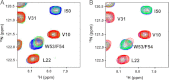
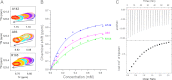
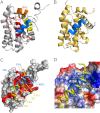
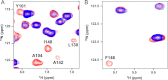
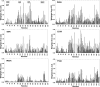
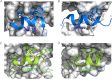
Molecular interactions between the tumor suppressor p53 and the anti-apoptotic Bcl-2 family proteins play an important role in the transcription-independent apoptosis of p53. The p53 transactivation domain (p53TAD) contains two conserved ΦXXΦΦ motifs (Φ indicates a bulky hydrophobic residue and X is any other residue) referred to as p53TAD1 (residues 15-29) and p53TAD2 (residues 39-57). We previously showed that p53TAD1 can act as a binding motif for anti-apoptotic Bcl-2 family proteins. In this study, we have identified p53TAD2 as a binding motif for anti-apoptotic Bcl-2 family proteins by using NMR spectroscopy, and we calculated the structures of Bcl-X(L)/Bcl-2 in complex with the p53TAD2 peptide. NMR chemical shift perturbation data showed that p53TAD2 peptide binds to diverse members of the anti-apoptotic Bcl-2 family independently of p53TAD1, and the binding between p53TAD2 and p53TAD1 to Bcl-X(L) is competitive. Refined structural models of the Bcl-X(L)·p53TAD2 and Bcl-2·p53TAD2 complexes showed that the binding sites occupied by p53TAD2 in Bcl-X(L) and Bcl-2 overlap well with those occupied by pro-apoptotic BH3 peptides. Taken together with the mutagenesis, isothermal titration calorimetry, and paramagnetic relaxation enhancement data, our structural comparisons provided the structural basis of p53TAD2-mediated interaction with the anti-apoptotic proteins, revealing that Bcl-X(L)/Bcl-2, MDM2, and cAMP-response element-binding protein-binding protein/p300 share highly similar modes of binding to the dual p53TAD motifs, p53TAD1 and p53TAD2. In conclusion, our results suggest that the dual-site interaction of p53TAD is a highly conserved mechanism underlying target protein binding in the transcription-dependent and transcription-independent apoptotic pathways of p53.
Figures


Similar articles
-
Structural basis for the conserved binding mechanism of MDM2-inhibiting peptides and anti-apoptotic Bcl-2 family proteins.Biochem Biophys Res Commun. 2014 Feb 28;445(1):120-5. doi: 10.1016/j.bbrc.2014.01.130. Epub 2014 Feb 1. Biochem Biophys Res Commun. 2014. PMID: 24491548
-
Targeting of p53 peptide analogues to anti-apoptotic Bcl-2 family proteins as revealed by NMR spectroscopy.Biochem Biophys Res Commun. 2014 Jan 17;443(3):882-7. doi: 10.1016/j.bbrc.2013.12.054. Epub 2013 Dec 14. Biochem Biophys Res Commun. 2014. PMID: 24342622
-
The MDM2-binding region in the transactivation domain of p53 also acts as a Bcl-X(L)-binding motif.Biochemistry. 2009 Dec 29;48(51):12159-68. doi: 10.1021/bi901188s. Biochemistry. 2009. PMID: 19916559
-
Structural biology of the Bcl-2 family of proteins.Biochim Biophys Acta. 2004 Mar 1;1644(2-3):83-94. doi: 10.1016/j.bbamcr.2003.08.012. Biochim Biophys Acta. 2004. PMID: 14996493 Review.
-
Discoveries and controversies in BCL-2 protein-mediated apoptosis.FEBS J. 2016 Jul;283(14):2690-700. doi: 10.1111/febs.13527. Epub 2015 Oct 27. FEBS J. 2016. PMID: 26411300 Review.
Cited by
-
Disruptive environmental chemicals and cellular mechanisms that confer resistance to cell death.Carcinogenesis. 2015 Jun;36 Suppl 1(Suppl 1):S89-110. doi: 10.1093/carcin/bgv032. Carcinogenesis. 2015. PMID: 26106145 Free PMC article. Review.
-
Investigation on tissue specific effects of pro-apoptotic micro RNAs revealed miR-147b as a potential biomarker in ovarian cancer prognosis.Oncotarget. 2017 Mar 21;8(12):18773-18791. doi: 10.18632/oncotarget.13095. Oncotarget. 2017. PMID: 27821806 Free PMC article.
-
The physical interaction of p53 and plakoglobin is necessary for their synergistic inhibition of migration and invasion.Oncotarget. 2016 May 3;7(18):26898-915. doi: 10.18632/oncotarget.8616. Oncotarget. 2016. PMID: 27058623 Free PMC article.
-
Characterization of the p300 Taz2-p53 TAD2 complex and comparison with the p300 Taz2-p53 TAD1 complex.Biochemistry. 2015 Mar 24;54(11):2001-10. doi: 10.1021/acs.biochem.5b00044. Epub 2015 Mar 16. Biochemistry. 2015. PMID: 25753752 Free PMC article.
-
Major apoptotic mechanisms and genes involved in apoptosis.Tumour Biol. 2016 Jul;37(7):8471-86. doi: 10.1007/s13277-016-5035-9. Epub 2016 Apr 9. Tumour Biol. 2016. PMID: 27059734 Review.
References
-
- Harris S. L., Levine A. J. (2005) The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–2908 - PubMed
-
- Sherr C. J. (2004) Principles of tumor suppression. Cell 116, 235–246 - PubMed
-
- Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 - PubMed
-
- Hollstein M., Sidransky D., Vogelstein B., Harris C. C. (1991) p53 mutations in human cancers. Science 253, 49–53 - PubMed
-
- Venot C., Maratrat M., Sierra V., Conseiller E., Debussche L. (1999) Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene 18, 2405–2410 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

