Cross-study homogeneity of psoriasis gene expression in skin across a large expression range
- PMID: 23308107
- PMCID: PMC3537625
- DOI: 10.1371/journal.pone.0052242
Cross-study homogeneity of psoriasis gene expression in skin across a large expression range
Abstract
Background: In psoriasis, only limited overlap between sets of genes identified as differentially expressed (psoriatic lesional vs. psoriatic non-lesional) was found using statistical and fold-change cut-offs. To provide a framework for utilizing prior psoriasis data sets we sought to understand the consistency of those sets.
Methodology/principal findings: Microarray expression profiling and qRT-PCR were used to characterize gene expression in PP and PN skin from psoriasis patients. cDNA (three new data sets) and cRNA hybridization (four existing data sets) data were compared using a common analysis pipeline. Agreement between data sets was assessed using varying qualitative and quantitative cut-offs to generate a DEG list in a source data set and then using other data sets to validate the list. Concordance increased from 67% across all probe sets to over 99% across more than 10,000 probe sets when statistical filters were employed. The fold-change behavior of individual genes tended to be consistent across the multiple data sets. We found that genes with <2-fold change values were quantitatively reproducible between pairs of data-sets. In a subset of transcripts with a role in inflammation changes detected by microarray were confirmed by qRT-PCR with high concordance. For transcripts with both PN and PP levels within the microarray dynamic range, microarray and qRT-PCR were quantitatively reproducible, including minimal fold-changes in IL13, TNFSF11, and TNFRSF11B and genes with >10-fold changes in either direction such as CHRM3, IL12B and IFNG.
Conclusions/significance: Gene expression changes in psoriatic lesions were consistent across different studies, despite differences in patient selection, sample handling, and microarray platforms but between-study comparisons showed stronger agreement within than between platforms. We could use cut-offs as low as log10(ratio) = 0.1 (fold-change = 1.26), generating larger gene lists that validate on independent data sets. The reproducibility of PP signatures across data sets suggests that different sample sets can be productively compared.
Conflict of interest statement
Figures
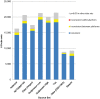


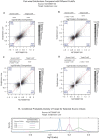
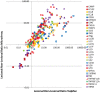
Similar articles
-
Reproducible Novel Transcriptional Differences Between Psoriatic Lesional and Non-Lesional Skin Show Increased Inflammation and Metabolism.J Drugs Dermatol. 2015 Aug;14(8):794-800. J Drugs Dermatol. 2015. PMID: 26267723
-
RNA-seq identifies a diminished differentiation gene signature in primary monolayer keratinocytes grown from lesional and uninvolved psoriatic skin.Sci Rep. 2017 Dec 22;7(1):18045. doi: 10.1038/s41598-017-18404-9. Sci Rep. 2017. PMID: 29273799 Free PMC article.
-
Global transcriptional analysis of psoriatic skin and blood confirms known disease-associated pathways and highlights novel genomic "hot spots" for differentially expressed genes.Genomics. 2012 Jul;100(1):18-26. doi: 10.1016/j.ygeno.2012.05.004. Epub 2012 May 11. Genomics. 2012. PMID: 22584065
-
MicroRNA profiling of psoriatic skin identifies 11 miRNAs associated with disease severity.Exp Dermatol. 2022 Apr;31(4):535-547. doi: 10.1111/exd.14497. Epub 2021 Nov 17. Exp Dermatol. 2022. PMID: 34748247 Review.
-
Transcriptional Basis of Psoriasis from Large Scale Gene Expression Studies: The Importance of Moving towards a Precision Medicine Approach.Int J Mol Sci. 2022 May 30;23(11):6130. doi: 10.3390/ijms23116130. Int J Mol Sci. 2022. PMID: 35682804 Free PMC article. Review.
Cited by
-
Upregulated SKP2 Empowers Epidermal Proliferation Through Downregulation of P27 Kip1.Ann Dermatol. 2024 Oct;36(5):282-291. doi: 10.5021/ad.23.118. Ann Dermatol. 2024. PMID: 39343755 Free PMC article.
-
Characterization of disease-specific cellular abundance profiles of chronic inflammatory skin conditions from deconvolution of biopsy samples.BMC Med Genomics. 2019 Aug 17;12(1):121. doi: 10.1186/s12920-019-0567-7. BMC Med Genomics. 2019. PMID: 31420038 Free PMC article.
-
Transcriptome Analysis Identifies Biomarkers for the Diagnosis and Management of Psoriasis Complicated with Depression.Clin Cosmet Investig Dermatol. 2023 May 18;16:1287-1301. doi: 10.2147/CCID.S413887. eCollection 2023. Clin Cosmet Investig Dermatol. 2023. PMID: 37223217 Free PMC article.
-
Lysosome Alterations in the Human Epithelial Cell Line HaCaT and Skin Specimens: Relevance to Psoriasis.Int J Mol Sci. 2019 May 7;20(9):2255. doi: 10.3390/ijms20092255. Int J Mol Sci. 2019. PMID: 31067781 Free PMC article.
-
Reproducibility enhancement and differential expression of non predefined functional gene sets in human genome.BMC Genomics. 2014 Dec 24;15(1):1181. doi: 10.1186/1471-2164-15-1181. BMC Genomics. 2014. PMID: 25539829 Free PMC article.
References
-
- Gottlieb AB, Grossman RM, Khandke L, Carter DM, Sehgal PB, et al. (1992) Studies of the effect of cyclosporine in psoriasis in vivo: combined effects on activated T lymphocytes and epidermal regenerative maturation. J Invest Dermatol 98: 302–309. - PubMed
-
- Guttman-Yassky E, Nograles KE, Krueger JG (2011) Contrasting pathogenesis of atopic dermatitis and psoriasis-Part I: Clinical and pathologic concepts. J Allergy Clin Immunol 127: 1110–1118. - PubMed
-
- Reischl J, Schwenke S, Beekman JM, Mrowietz U, Stürzebecher S, et al. (2007) Increased expression of Wnt5a in psoriatic plaques. J Invest Dermatol 127: 163–169. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

