PIK3CA genotype and a PIK3CA mutation-related gene signature and response to everolimus and letrozole in estrogen receptor positive breast cancer
- PMID: 23301057
- PMCID: PMC3534682
- DOI: 10.1371/journal.pone.0053292
PIK3CA genotype and a PIK3CA mutation-related gene signature and response to everolimus and letrozole in estrogen receptor positive breast cancer
Erratum in
-
Correction: PIK3CA Genotype and a PIK3CA Mutation-Related Gene Signature and Response to Everolimus and Letrozole in Estrogen Receptor Positive Breast Cancer.PLoS One. 2019 Apr 23;14(4):e0216175. doi: 10.1371/journal.pone.0216175. eCollection 2019. PLoS One. 2019. PMID: 31013327 Free PMC article.
Abstract
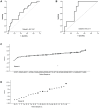
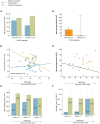
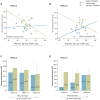
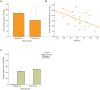
The phosphatidylinositol 3' kinase (PI3K) pathway is commonly activated in breast cancer and aberrations such as PI3K mutations are common. Recent exciting clinical trial results in advanced estrogen receptor-positive (ER) breast cancer support mTOR activation is a major means of estrogen-independent tumor growth. Hence the means to identify a responsive breast cancer population that would most benefit from these compounds in the adjuvant or earlier stage setting is of high interest. Here we study PIK3CA genotype as well as a previously reported PI3K/mTOR-pathway gene signature (PIK3CA-GS) and their ability to estimate the level of PI3K pathway activation in two clinical trials of newly diagnosed ER-positive breast cancer patients- a total of 81 patients- one of which was randomized between letrozole and placebo vs letrozole and everolimus. The main objectives were to correlate the baseline PIK3CA genotype and GS with the relative change from baseline to day 15 in Ki67 (which has been shown to be prognostic in breast cancer) and phosphorylated S6 (S240) immunohistochemistry (a substrate of mTOR). In the randomized dataset, the PIK3CA-GS could identify those patients with the largest relative decreases in Ki67 to letrozole/everolimus (R = -0.43, p = 0.008) compared with letrozole/placebo (R = 0.07, p = 0.58; interaction test p = 0.02). In a second dataset of pre-surgical everolimus alone, the PIK3CA-GS was not significantly correlated with relative change in Ki67 (R = -0.11, p = 0.37) but with relative change in phosphorlyated S6 (S240) (R = -0.46, p = 0.028). PIK3CA genotype was not significantly associated with any endpoint in either datasets. Our results suggest that the PIK3CA-GS has potential to identify those ER-positive BCs who may benefit from the addition of everolimus to letrozole. Further evaluation of the PIK3CA-GS as a predictive biomarker is warranted as it may facilitate better selection of responsive patient populations for mTOR inhibition in combination with letrozole.
Conflict of interest statement
Figures
Similar articles
-
Relationship of PIK3CA mutation and pathway activity with antiproliferative response to aromatase inhibition.Breast Cancer Res. 2014 Jun 30;16(3):R68. doi: 10.1186/bcr3683. Breast Cancer Res. 2014. PMID: 24981670 Free PMC article. Clinical Trial.
-
Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer.J Clin Oncol. 2009 Jun 1;27(16):2630-7. doi: 10.1200/JCO.2008.18.8391. Epub 2009 Apr 20. J Clin Oncol. 2009. PMID: 19380449 Clinical Trial.
-
Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer.Breast Cancer Res. 2011 Mar 1;13(2):R21. doi: 10.1186/bcr2833. Breast Cancer Res. 2011. PMID: 21362200 Free PMC article.
-
Co-targeting estrogen receptor and HER2 pathways in breast cancer.Breast. 2014 Feb;23(1):2-9. doi: 10.1016/j.breast.2013.09.006. Epub 2013 Oct 28. Breast. 2014. PMID: 24176518 Review.
-
Future directions in the treatment of hormone-sensitive advanced breast cancer: the RAD001 (Everolimus)-letrozole clinical program.Semin Oncol. 2006 Apr;33(2 Suppl 7):S18-25. doi: 10.1053/j.seminoncol.2006.03.024. Semin Oncol. 2006. PMID: 16730273 Review.
Cited by
-
Molecular Biomarker Expression in Window of Opportunity Studies for Oestrogen Receptor Positive Breast Cancer-A Systematic Review of the Literature.Cancers (Basel). 2022 Oct 14;14(20):5027. doi: 10.3390/cancers14205027. Cancers (Basel). 2022. PMID: 36291809 Free PMC article. Review.
-
Relationship of PIK3CA mutation and pathway activity with antiproliferative response to aromatase inhibition.Breast Cancer Res. 2014 Jun 30;16(3):R68. doi: 10.1186/bcr3683. Breast Cancer Res. 2014. PMID: 24981670 Free PMC article. Clinical Trial.
-
BAP1-related signature predicts benefits from immunotherapy over VEGFR/mTOR inhibitors in ccRCC: a retrospective analysis of JAVELIN Renal 101 and checkmate-009/010/025 trials.Cancer Immunol Immunother. 2023 Aug;72(8):2557-2572. doi: 10.1007/s00262-023-03424-4. Epub 2023 Apr 12. Cancer Immunol Immunother. 2023. PMID: 37046008 Free PMC article.
-
Leveraging a pharmacogenomics knowledgebase to formulate a drug response phenotype terminology for genomic medicine.Bioinformatics. 2022 Nov 30;38(23):5279-5287. doi: 10.1093/bioinformatics/btac646. Bioinformatics. 2022. PMID: 36222570 Free PMC article.
-
Biomarkers of response and resistance to PI3K inhibitors in estrogen receptor-positive breast cancer patients and combination therapies involving PI3K inhibitors.Ann Oncol. 2019 Dec 1;30(Suppl_10):x27-x42. doi: 10.1093/annonc/mdz280. Ann Oncol. 2019. PMID: 31859350 Free PMC article. Review.
References
-
- Engelman JA (2009) Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 9: 550–562. - PubMed
-
- O'Brien C, Wallin JJ, Sampath D, GuhaThakurta D, Savage H, et al. (2010) Predictive biomarkers of sensitivity to the phosphatidylinositol 3' kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin Cancer Res 16: 3670–3683. - PubMed
-
- Boulay A, Rudloff J, Ye J, Zumstein-Mecker S, O'Reilly T, et al. (2005) Dual inhibition of mTOR and estrogen receptor signaling in vitro induces cell death in models of breast cancer. Clin Cancer Res 11: 5319–5328. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

