Atomic scissors: a new method of tracking the 5-bromo-2'-deoxyuridine-labeled DNA in situ
- PMID: 23300711
- PMCID: PMC3530445
- DOI: 10.1371/journal.pone.0052584
Atomic scissors: a new method of tracking the 5-bromo-2'-deoxyuridine-labeled DNA in situ
Abstract
A new method of the light microscopy detection of BrdU-labeled DNA in situ is described. It is based on the oxidative attack at the deoxyribose moiety by copper(I) in the presence of oxygen, which leads to the abstraction of hydrogen atom from deoxyribose culminating in the elimination of the nucleobase, scission of the nucleic-acid strand and formation of frequent gaps. The gaps allow the reaction of the antibodies with the commonly used markers of replication (e.g. 5-bromo-2'-deoxyuridine), which are otherwise masked. The method developed makes it possible to detect nuclear and mitochondrial DNA replication efficiently. In most cases, it does not inhibit effective protein detections and in addition enables simultaneous localization of newly-synthesized RNA. The alternative presently-used methods result in protein denaturation and/or extensive DNA cleavage followed by the DNA-bound proteins peeling off.
Conflict of interest statement
Figures

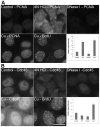
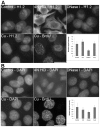
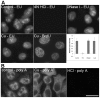
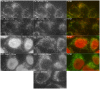
Similar articles
-
A New Method of the Visualization of the Double-Stranded Mitochondrial and Nuclear DNA.PLoS One. 2013 Jun 18;8(6):e66864. doi: 10.1371/journal.pone.0066864. Print 2013. PLoS One. 2013. PMID: 23825578 Free PMC article.
-
Tracking Mitochondrial DNA In Situ.Methods Mol Biol. 2016;1351:81-92. doi: 10.1007/978-1-4939-3040-1_7. Methods Mol Biol. 2016. PMID: 26530676
-
From "Click" to "Fenton" chemistry for 5-bromo-2'-deoxyuridine determination.Cytometry A. 2013 Nov;83(11):989-1000. doi: 10.1002/cyto.a.22343. Epub 2013 Aug 13. Cytometry A. 2013. PMID: 23943293
-
Release of viruses and viral DNA from nucleus to cytoplasm of HeLa cells at late stages of productive adenovirus infection as revealed by electron microscope in situ hybridization.Biol Cell. 1998 Jan;90(1):5-38. doi: 10.1016/s0248-4900(98)80230-x. Biol Cell. 1998. PMID: 9691424 Review.
-
Thymidine analogues for tracking DNA synthesis.Molecules. 2011 Sep 15;16(9):7980-93. doi: 10.3390/molecules16097980. Molecules. 2011. PMID: 21921870 Free PMC article. Review.
Cited by
-
Dr Jekyll and Mr Hyde: a strange case of 5-ethynyl-2'-deoxyuridine and 5-ethynyl-2'-deoxycytidine.Open Biol. 2016 Jan;6(1):150172. doi: 10.1098/rsob.150172. Open Biol. 2016. PMID: 26740587 Free PMC article.
-
Analysis of a temperature-sensitive mutation in Uba1: Effects of the click reaction on subsequent immunolabeling of proteins involved in DNA replication.FEBS Open Bio. 2015 Mar 3;5:167-74. doi: 10.1016/j.fob.2015.02.004. eCollection 2015. FEBS Open Bio. 2015. PMID: 25834782 Free PMC article.
-
A New Method of the Visualization of the Double-Stranded Mitochondrial and Nuclear DNA.PLoS One. 2013 Jun 18;8(6):e66864. doi: 10.1371/journal.pone.0066864. Print 2013. PLoS One. 2013. PMID: 23825578 Free PMC article.
-
A new technique for the analysis of metabolic pathways of cytidine analogues and cytidine deaminase activities in cells.Sci Rep. 2023 Nov 22;13(1):20530. doi: 10.1038/s41598-023-47792-4. Sci Rep. 2023. PMID: 37993628 Free PMC article.
-
Quantification of fixed adherent cells using a strong enhancer of the fluorescence of DNA dyes.Sci Rep. 2019 Jun 18;9(1):8701. doi: 10.1038/s41598-019-45217-9. Sci Rep. 2019. PMID: 31213648 Free PMC article.
References
-
- Dimitrova DS, Berezney R (2002) The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J Cell Sci 115: 4037–4051. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

