Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities
- PMID: 23297216
- PMCID: PMC3557073
- DOI: 10.1073/pnas.1218509110
Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities
Abstract
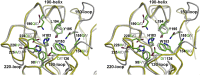
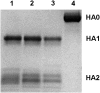
Bat influenza virus H17N10 represents a distinct lineage of influenza A viruses with gene segments coding for proteins that are homologs of the surface antigens, hemagglutinin (HA) and neuraminidase (NA). Our recent study of the N10 NA homolog revealed an NA-like structure, but with a highly divergent putative active site exhibiting little or no NA activity, and provided strong motivation for performing equivalent structural and functional analyses of the H17 HA protein. The overall structure of the H17 HA homolog from A/little yellow-shouldered bat/Guatemala/060/2010 at 3.18 Å resolution is very similar to other influenza HAs, with a putative receptor-binding site containing some conserved aromatic residues that form the base of the sialic acid binding site. However, the rest of the H17 receptor-binding site differs substantially from the other HA subtypes, including substitution of other conserved residues associated with receptor binding. Significantly, electrostatic potential analyses reveal that this putative receptor-binding site is highly acidic, making it unfavorable to bind any negatively charged sialylated receptors, consistent with the recombinant H17 protein exhibiting no detectable binding to sialylated glycans. Furthermore, the fusion mechanism is also distinct; trypsin digestion with recombinant H17 protein, when exposed to pH 4.0, did not degrade the HA1 and HA2, in contrast to other HAs. These distinct structural features and functional differences suggest that the H17 HA behaves very differently compared with other influenza HAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism.Cell Rep. 2013 Mar 28;3(3):769-78. doi: 10.1016/j.celrep.2013.01.025. Epub 2013 Feb 21. Cell Rep. 2013. PMID: 23434510
-
Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18903-8. doi: 10.1073/pnas.1212579109. Epub 2012 Sep 24. Proc Natl Acad Sci U S A. 2012. PMID: 23012478 Free PMC article.
-
Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18897-902. doi: 10.1073/pnas.1211037109. Epub 2012 Sep 24. Proc Natl Acad Sci U S A. 2012. PMID: 23012237 Free PMC article.
-
Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin.Annu Rev Biochem. 2000;69:531-69. doi: 10.1146/annurev.biochem.69.1.531. Annu Rev Biochem. 2000. PMID: 10966468 Review.
-
[Structure and function of the hemagglutinin of influenza viruses].Nihon Rinsho. 1997 Oct;55(10):2562-9. Nihon Rinsho. 1997. PMID: 9360372 Review. Japanese.
Cited by
-
Influenza Hemagglutinin Structures and Antibody Recognition.Cold Spring Harb Perspect Med. 2020 Aug 3;10(8):a038778. doi: 10.1101/cshperspect.a038778. Cold Spring Harb Perspect Med. 2020. PMID: 31871236 Free PMC article. Review.
-
MHC class II proteins mediate cross-species entry of bat influenza viruses.Nature. 2019 Mar;567(7746):109-112. doi: 10.1038/s41586-019-0955-3. Epub 2019 Feb 20. Nature. 2019. PMID: 30787439
-
Recent advances in "universal" influenza virus antibodies: the rise of a hidden trimeric interface in hemagglutinin globular head.Front Med. 2020 Apr;14(2):149-159. doi: 10.1007/s11684-020-0764-y. Epub 2020 Apr 1. Front Med. 2020. PMID: 32239416 Free PMC article. Review.
-
Characterization of an Avian Influenza Virus H9N2 Strain Isolated from Dove in Southern China.Genome Announc. 2018 May 3;6(18):e00369-18. doi: 10.1128/genomeA.00369-18. Genome Announc. 2018. PMID: 29724842 Free PMC article.
-
Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus.Nat Commun. 2014 Apr 10;5:3614. doi: 10.1038/ncomms4614. Nat Commun. 2014. PMID: 24717798 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

