Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity
- PMID: 23293005
- PMCID: PMC3575835
- DOI: 10.1093/nar/gks1205
Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity
Abstract
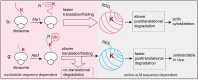
Messenger RNA is a key component of an intricate regulatory network of its own. It accommodates numerous nucleotide signals that overlap protein coding sequences and are responsible for multiple levels of regulation and generation of biological complexity. A wealth of structural and regulatory information, which mRNA carries in addition to the encoded amino acid sequence, raises the question of how these signals and overlapping codes are delineated along non-synonymous and synonymous positions in protein coding regions, especially in eukaryotes. Silent or synonymous codon positions, which do not determine amino acid sequences of the encoded proteins, define mRNA secondary structure and stability and affect the rate of translation, folding and post-translational modifications of nascent polypeptides. The RNA level selection is acting on synonymous sites in both prokaryotes and eukaryotes and is more common than previously thought. Selection pressure on the coding gene regions follows three-nucleotide periodic pattern of nucleotide base-pairing in mRNA, which is imposed by the genetic code. Synonymous positions of the coding regions have a higher level of hybridization potential relative to non-synonymous positions, and are multifunctional in their regulatory and structural roles. Recent experimental evidence and analysis of mRNA structure and interspecies conservation suggest that there is an evolutionary tradeoff between selective pressure acting at the RNA and protein levels. Here we provide a comprehensive overview of the studies that define the role of silent positions in regulating RNA structure and processing that exert downstream effects on proteins and their functions.
Figures

Similar articles
-
Selection on synonymous codons in mammalian rhodopsins: a possible role in optimizing translational processes.BMC Evol Biol. 2014 May 3;14:96. doi: 10.1186/1471-2148-14-96. BMC Evol Biol. 2014. PMID: 24884412 Free PMC article.
-
Synonymous nucleotide modification of the KCNH2 gene affects both mRNA characteristics and translation of the encoded hERG ion channel.J Biol Chem. 2018 Aug 3;293(31):12120-12136. doi: 10.1074/jbc.RA118.001805. Epub 2018 Jun 15. J Biol Chem. 2018. PMID: 29907571 Free PMC article.
-
Roles for Synonymous Codon Usage in Protein Biogenesis.Annu Rev Biophys. 2015;44:143-66. doi: 10.1146/annurev-biophys-060414-034333. Epub 2015 Feb 26. Annu Rev Biophys. 2015. PMID: 25747594 Review.
-
Decoding mechanisms by which silent codon changes influence protein biogenesis and function.Int J Biochem Cell Biol. 2015 Jul;64:58-74. doi: 10.1016/j.biocel.2015.03.011. Epub 2015 Mar 26. Int J Biochem Cell Biol. 2015. PMID: 25817479 Free PMC article. Review.
-
Molecular Mechanisms and the Significance of Synonymous Mutations.Biomolecules. 2024 Jan 20;14(1):132. doi: 10.3390/biom14010132. Biomolecules. 2024. PMID: 38275761 Free PMC article. Review.
Cited by
-
Role of mRNA structure in the control of protein folding.Nucleic Acids Res. 2016 Dec 15;44(22):10898-10911. doi: 10.1093/nar/gkw671. Epub 2016 Jul 27. Nucleic Acids Res. 2016. PMID: 27466388 Free PMC article.
-
Analysis of codon usage patterns in Ginkgo biloba reveals codon usage tendency from A/U-ending to G/C-ending.Sci Rep. 2016 Nov 3;6:35927. doi: 10.1038/srep35927. Sci Rep. 2016. PMID: 27808241 Free PMC article.
-
Systematic errors in annotations of truncations, loss-of-function and synonymous variants.Front Genet. 2023 Jan 13;14:1015017. doi: 10.3389/fgene.2023.1015017. eCollection 2023. Front Genet. 2023. PMID: 36713076 Free PMC article.
-
Intronic RNA: Ad'junk' mediator of post-transcriptional gene regulation.Biochim Biophys Acta Gene Regul Mech. 2019 Nov-Dec;1862(11-12):194439. doi: 10.1016/j.bbagrm.2019.194439. Epub 2019 Nov 1. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31682938 Free PMC article. Review.
-
Strategies and Patterns of Codon Bias in Molluscum Contagiosum Virus.Pathogens. 2021 Dec 20;10(12):1649. doi: 10.3390/pathogens10121649. Pathogens. 2021. PMID: 34959603 Free PMC article.
References
-
- Claverie JM. Gene number. What if there are only 30,000 human genes? Science. 2001;291:1255–1257. - PubMed
-
- Pennisi E. Human genome. A low number wins the GeneSweep Pool. Science. 2003;300:1484. - PubMed
-
- Stein LD. Human genome: end of the beginning. Nature. 2004;431:915–916. - PubMed
-
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

