Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa)
- PMID: 23264228
- PMCID: PMC3682022
- DOI: 10.1007/s12192-012-0395-6
Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa)
Abstract
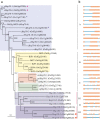
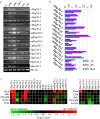
Heat stress results in misfolding and aggregation of cellular proteins. Heat shock proteins (Hsp) enable the cells to maintain proper folding of proteins, both in unstressed as well as stressed conditions. Hsp70 genes encode for a group of highly conserved chaperone proteins across the living systems encompassing bacteria, plants, and animals. In the cellular chaperone network, Hsp70 family proteins interconnect other chaperones and play a dominant role in various cell processes. To assess the functionality of rice Hsp70 genes, rice genome database was analyzed. Rice genome contains 32 Hsp70 genes. Rice Hsp70 superfamily genes are represented by 24 Hsp70 family and 8 Hsp110 family members. Promoter and transcript expression analysis divulges that Hsp70 superfamily genes plays important role in heat stress. Ssc1 (mitochondrial Hsp70 protein in yeast) deleted yeast show compromised growth at 37 °C. Three mitochondrial rice Hsp70 sequences (i.e., mtHsp70-1, mtHsp70-2, and mtHsp70-3) complemented the Ssc1 mutation of yeast to differential extents. The information presented in this study provides detailed understanding of the Hsp70 protein family of rice, the crop species that is the major food for the world population.
Figures


Similar articles
-
Genome-wide identification of heat shock proteins (Hsps) and Hsp interactors in rice: Hsp70s as a case study.BMC Genomics. 2014 May 7;15(1):344. doi: 10.1186/1471-2164-15-344. BMC Genomics. 2014. PMID: 24884676 Free PMC article.
-
In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70.PLoS One. 2010 Sep 15;5(9):e12761. doi: 10.1371/journal.pone.0012761. PLoS One. 2010. PMID: 20856808 Free PMC article.
-
A genomewide analysis of genes for the heat shock protein 70 chaperone system in the ascidian Ciona intestinalis.Cell Stress Chaperones. 2006 Spring;11(1):23-33. doi: 10.1379/csc-137r.1. Cell Stress Chaperones. 2006. PMID: 16572726 Free PMC article.
-
An Hsp70 family chaperone, mortalin/mthsp70/PBP74/Grp75: what, when, and where?Cell Stress Chaperones. 2002 Jul;7(3):309-16. doi: 10.1379/1466-1268(2002)007<0309:ahfcmm>2.0.co;2. Cell Stress Chaperones. 2002. PMID: 12482206 Free PMC article. Review.
-
Mitochondrial HSP70 Chaperone System-The Influence of Post-Translational Modifications and Involvement in Human Diseases.Int J Mol Sci. 2021 Jul 28;22(15):8077. doi: 10.3390/ijms22158077. Int J Mol Sci. 2021. PMID: 34360841 Free PMC article. Review.
Cited by
-
Sequence analysis of the Hsp70 family in moss and evaluation of their functions in abiotic stress responses.Sci Rep. 2016 Sep 20;6:33650. doi: 10.1038/srep33650. Sci Rep. 2016. PMID: 27644410 Free PMC article.
-
Identifying Heat Shock Protein Families from Imbalanced Data by Using Combined Features.Comput Math Methods Med. 2020 Sep 23;2020:8894478. doi: 10.1155/2020/8894478. eCollection 2020. Comput Math Methods Med. 2020. PMID: 33029195 Free PMC article.
-
Identification of watermelon heat shock protein members and tissue-specific gene expression analysis under combined drought and heat stresses.Turk J Biol. 2019 Dec 13;43(4):404-419. doi: 10.3906/biy-1907-5. eCollection 2019. Turk J Biol. 2019. PMID: 31892809 Free PMC article.
-
Evolutionary analysis and expression profiling of the HSP70 gene family in response to abiotic stresses in tomato (Solanum lycopersicum).Sci Prog. 2023 Jan-Mar;106(1):368504221148843. doi: 10.1177/00368504221148843. Sci Prog. 2023. PMID: 36650980 Free PMC article.
-
Regulation of heat shock proteins 70 and their role in plant immunity.J Exp Bot. 2022 Apr 5;73(7):1894-1909. doi: 10.1093/jxb/erab549. J Exp Bot. 2022. PMID: 35022724 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

