Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes
- PMID: 23259493
- PMCID: PMC3570280
- DOI: 10.1186/1741-7007-10-107
Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes
Abstract
Background: Calcium-activated photoproteins are luciferase variants found in photocyte cells of bioluminescent jellyfish (Phylum Cnidaria) and comb jellies (Phylum Ctenophora). The complete genomic sequence from the ctenophore Mnemiopsis leidyi, a representative of the earliest branch of animals that emit light, provided an opportunity to examine the genome of an organism that uses this class of luciferase for bioluminescence and to look for genes involved in light reception. To determine when photoprotein genes first arose, we examined the genomic sequence from other early-branching taxa. We combined our genomic survey with gene trees, developmental expression patterns, and functional protein assays of photoproteins and opsins to provide a comprehensive view of light production and light reception in Mnemiopsis.
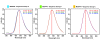
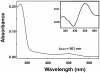
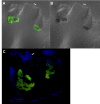
Results: The Mnemiopsis genome has 10 full-length photoprotein genes situated within two genomic clusters with high sequence conservation that are maintained due to strong purifying selection and concerted evolution. Photoprotein-like genes were also identified in the genomes of the non-luminescent sponge Amphimedon queenslandica and the non-luminescent cnidarian Nematostella vectensis, and phylogenomic analysis demonstrated that photoprotein genes arose at the base of all animals. Photoprotein gene expression in Mnemiopsis embryos begins during gastrulation in migrating precursors to photocytes and persists throughout development in the canals where photocytes reside. We identified three putative opsin genes in the Mnemiopsis genome and show that they do not group with well-known bilaterian opsin subfamilies. Interestingly, photoprotein transcripts are co-expressed with two of the putative opsins in developing photocytes. Opsin expression is also seen in the apical sensory organ. We present evidence that one opsin functions as a photopigment in vitro, absorbing light at wavelengths that overlap with peak photoprotein light emission, raising the hypothesis that light production and light reception may be functionally connected in ctenophore photocytes. We also present genomic evidence of a complete ciliary phototransduction cascade in Mnemiopsis.
Conclusions: This study elucidates the genomic organization, evolutionary history, and developmental expression of photoprotein and opsin genes in the ctenophore Mnemiopsis leidyi, introduces a novel dual role for ctenophore photocytes in both bioluminescence and phototransduction, and raises the possibility that light production and light reception are linked in this early-branching non-bilaterian animal.
Figures


Similar articles
-
The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution.Science. 2013 Dec 13;342(6164):1242592. doi: 10.1126/science.1242592. Science. 2013. PMID: 24337300 Free PMC article.
-
Evolution of the TGF-β signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi.PLoS One. 2011;6(9):e24152. doi: 10.1371/journal.pone.0024152. Epub 2011 Sep 8. PLoS One. 2011. PMID: 21931657 Free PMC article.
-
Expression and characterization of the calcium-activated photoprotein from the ctenophore Bathocyroe fosteri: insights into light-sensitive photoproteins.Biochem Biophys Res Commun. 2013 Feb 8;431(2):360-6. doi: 10.1016/j.bbrc.2012.12.026. Epub 2012 Dec 19. Biochem Biophys Res Commun. 2013. PMID: 23262181 Free PMC article.
-
Whole-Body Regeneration in the Lobate Ctenophore Mnemiopsis leidyi.Genes (Basel). 2021 Jun 5;12(6):867. doi: 10.3390/genes12060867. Genes (Basel). 2021. PMID: 34198839 Free PMC article. Review.
-
Shedding new light on opsin evolution.Proc Biol Sci. 2012 Jan 7;279(1726):3-14. doi: 10.1098/rspb.2011.1819. Epub 2011 Oct 19. Proc Biol Sci. 2012. PMID: 22012981 Free PMC article. Review.
Cited by
-
Convergent evolution of neural systems in ctenophores.J Exp Biol. 2015 Feb 15;218(Pt 4):598-611. doi: 10.1242/jeb.110692. J Exp Biol. 2015. PMID: 25696823 Free PMC article. Review.
-
Transcriptional remodelling upon light removal in a model cnidarian: Losses and gains in gene expression.Mol Ecol. 2019 Jul;28(14):3413-3426. doi: 10.1111/mec.15163. Epub 2019 Jul 15. Mol Ecol. 2019. PMID: 31264275 Free PMC article.
-
A new transcriptome and transcriptome profiling of adult and larval tissue in the box jellyfish Alatina alata: an emerging model for studying venom, vision and sex.BMC Genomics. 2016 Aug 17;17:650. doi: 10.1186/s12864-016-2944-3. BMC Genomics. 2016. PMID: 27535656 Free PMC article.
-
The ctenophore Mnemiopsis leidyi deploys a rapid injury response dating back to the last common animal ancestor.Commun Biol. 2024 Feb 19;7(1):203. doi: 10.1038/s42003-024-05901-7. Commun Biol. 2024. PMID: 38374160 Free PMC article.
-
Molecular evolution and expression of opsin genes in Hydra vulgaris.BMC Genomics. 2019 Dec 17;20(1):992. doi: 10.1186/s12864-019-6349-y. BMC Genomics. 2019. PMID: 31847811 Free PMC article.
References
-
- Harper RD, Case JF. Disruptive counterillumination and its anti-predatory value in the plainfish midshipman Porichthys notatus. Marine Biology. 1999;134:529–540. doi: 10.1007/s002270050568. - DOI
-
- Frank TM, Widder EA, Latz MI, Case JF. Dietary maintenance of bioluminescence in a deep-sea mysid. J Exp Biol. 1984;109:385–389.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

