MicroRNAs and essential components of the microRNA processing machinery are not encoded in the genome of the ctenophore Mnemiopsis leidyi
- PMID: 23256903
- PMCID: PMC3563456
- DOI: 10.1186/1471-2164-13-714
MicroRNAs and essential components of the microRNA processing machinery are not encoded in the genome of the ctenophore Mnemiopsis leidyi
Abstract
Background: MicroRNAs play a vital role in the regulation of gene expression and have been identified in every animal with a sequenced genome examined thus far, except for the placozoan Trichoplax. The genomic repertoires of metazoan microRNAs have become increasingly endorsed as phylogenetic characters and drivers of biological complexity.
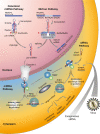
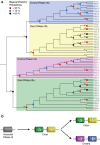
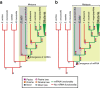
Results: In this study, we report the first investigation of microRNAs in a species from the phylum Ctenophora. We use short RNA sequencing and the assembled genome of the lobate ctenophore Mnemiopsis leidyi to show that this species appears to lack any recognizable microRNAs, as well as the nuclear proteins Drosha and Pasha, which are critical to canonical microRNA biogenesis. This finding represents the first reported case of a metazoan lacking a Drosha protein.
Conclusions: Recent phylogenomic analyses suggest that Mnemiopsis may be the earliest branching metazoan lineage. If this is true, then the origins of canonical microRNA biogenesis and microRNA-mediated gene regulation may postdate the last common metazoan ancestor. Alternatively, canonical microRNA functionality may have been lost independently in the lineages leading to both Mnemiopsis and the placozoan Trichoplax, suggesting that microRNA functionality was not critical until much later in metazoan evolution.
Figures

Similar articles
-
Multigenerational laboratory culture of pelagic ctenophores and CRISPR-Cas9 genome editing in the lobate Mnemiopsis leidyi.Nat Protoc. 2022 Aug;17(8):1868-1900. doi: 10.1038/s41596-022-00702-w. Epub 2022 Jun 13. Nat Protoc. 2022. PMID: 35697825 Review.
-
Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes.BMC Biol. 2012 Dec 21;10:107. doi: 10.1186/1741-7007-10-107. BMC Biol. 2012. PMID: 23259493 Free PMC article.
-
Evolution of the TGF-β signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi.PLoS One. 2011;6(9):e24152. doi: 10.1371/journal.pone.0024152. Epub 2011 Sep 8. PLoS One. 2011. PMID: 21931657 Free PMC article.
-
Nuclear receptors from the ctenophore Mnemiopsis leidyi lack a zinc-finger DNA-binding domain: lineage-specific loss or ancestral condition in the emergence of the nuclear receptor superfamily?Evodevo. 2011 Feb 3;2(1):3. doi: 10.1186/2041-9139-2-3. Evodevo. 2011. PMID: 21291545 Free PMC article.
-
Whole-Body Regeneration in the Lobate Ctenophore Mnemiopsis leidyi.Genes (Basel). 2021 Jun 5;12(6):867. doi: 10.3390/genes12060867. Genes (Basel). 2021. PMID: 34198839 Free PMC article. Review.
Cited by
-
KLF/SP Transcription Factor Family Evolution: Expansion, Diversification, and Innovation in Eukaryotes.Genome Biol Evol. 2015 Jul 30;7(8):2289-309. doi: 10.1093/gbe/evv141. Genome Biol Evol. 2015. PMID: 26232396 Free PMC article.
-
Multigenerational laboratory culture of pelagic ctenophores and CRISPR-Cas9 genome editing in the lobate Mnemiopsis leidyi.Nat Protoc. 2022 Aug;17(8):1868-1900. doi: 10.1038/s41596-022-00702-w. Epub 2022 Jun 13. Nat Protoc. 2022. PMID: 35697825 Review.
-
Functional characterization of a 'plant-like' HYL1 homolog in the cnidarian Nematostella vectensis indicates a conserved involvement in microRNA biogenesis.Elife. 2022 Mar 15;11:e69464. doi: 10.7554/eLife.69464. Elife. 2022. PMID: 35289745 Free PMC article.
-
Diverse RNA interference strategies in early-branching metazoans.BMC Evol Biol. 2018 Nov 1;18(1):160. doi: 10.1186/s12862-018-1274-2. BMC Evol Biol. 2018. PMID: 30382896 Free PMC article.
-
Regulatory logic and transposable element dynamics in nematode worm genomes.bioRxiv [Preprint]. 2024 Sep 16:2024.09.15.613132. doi: 10.1101/2024.09.15.613132. bioRxiv. 2024. PMID: 39345564 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

