A short cis-acting motif in the M112-113 promoter region is essential for IE3 to activate M112-113 gene expression and is important for murine cytomegalovirus replication
- PMID: 23255797
- PMCID: PMC3571397
- DOI: 10.1128/JVI.03171-12
A short cis-acting motif in the M112-113 promoter region is essential for IE3 to activate M112-113 gene expression and is important for murine cytomegalovirus replication
Erratum in
-
Correction for Perez et al., "A Short cis-Acting Motif in the M112-113 Promoter Region Is Essential for IE3 To Activate M112-113 Gene Expression and Is Important for Murine Cytomegalovirus Replication".J Virol. 2017 Nov 14;91(23):e01527-17. doi: 10.1128/JVI.01527-17. Print 2017 Dec 1. J Virol. 2017. PMID: 29138332 Free PMC article. No abstract available.
Abstract
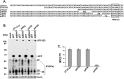
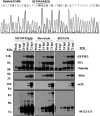
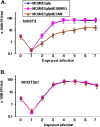
Immediate-early 3 (IE3) gene products are required to activate early (E)-stage gene expression of murine cytomegaloviruses (MCMV). The first early gene activated by IE3 is the M112-113 gene (also called E1), although a complete understanding of the activation mechanism is still lacking. In this paper, we identify a 10-bp cis-regulating motif upstream of the M112-113 TATA box as important for IE3 activation of M112-113 expression. Results from DNA affinity assays and chromatin immunoprecipitation assays show that the association of IE3 with the M112-113 gene promoter was eliminated by deletion of the 10-bp DNA sequence, now named IE3AM (for IE3 activating motif). In addition, IE3 interacts with TATA box binding protein (TBP), a core protein of TFIID (transcription initiation) complexes. Finally, we created an IE3AM-deleted MCMV (MCMVdIE3AM) using a bacterial artificial chromosome system. The mutant virus can still replicate in NIH 3T3 cells but at a significantly lower level. The defectiveness of the MCMVdIE3AM infection can be rescued in an M112-113-complemented cell line. Our results suggest that the interactions of IE3 with IE3AM and with TBP stabilize the TFIID complex at the M112-113 promoter such that M112-113 gene expression can be activated and/or enhanced.
Figures


Similar articles
-
The Canonical Immediate Early 3 Gene Product pIE611 of Mouse Cytomegalovirus Is Dispensable for Viral Replication but Mediates Transcriptional and Posttranscriptional Regulation of Viral Gene Products.J Virol. 2015 Aug;89(16):8590-8. doi: 10.1128/JVI.01234-15. Epub 2015 Jun 10. J Virol. 2015. PMID: 26063418 Free PMC article.
-
One of the Triple Poly(A) Signals in the M112-113 Gene Is Important and Sufficient for Stabilizing the M112-113 mRNA and the Replication of Murine Cytomegalovirus.Viruses. 2020 Aug 28;12(9):954. doi: 10.3390/v12090954. Viruses. 2020. PMID: 32872150 Free PMC article.
-
Mouse cytomegalovirus early M112/113 proteins control the repressive effect of IE3 on the major immediate-early promoter.J Virol. 2005 Jan;79(1):257-63. doi: 10.1128/JVI.79.1.257-263.2005. J Virol. 2005. PMID: 15596821 Free PMC article.
-
The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth.J Virol. 2000 Dec;74(23):11129-36. doi: 10.1128/jvi.74.23.11129-11136.2000. J Virol. 2000. PMID: 11070009 Free PMC article.
-
Murine cytomegalovirus major immediate-early protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation.J Gen Virol. 2010 Nov;91(Pt 11):2664-76. doi: 10.1099/vir.0.022301-0. Epub 2010 Jul 14. J Gen Virol. 2010. PMID: 20631086 Free PMC article.
Cited by
-
Seminal plasma and semen amyloids enhance cytomegalovirus infection in cell culture.J Virol. 2013 Dec;87(23):12583-91. doi: 10.1128/JVI.02083-13. Epub 2013 Sep 11. J Virol. 2013. PMID: 24027327 Free PMC article.
-
Molecular investigation of the 7.2 kb RNA of murine cytomegalovirus.Virol J. 2013 Dec 2;10:348. doi: 10.1186/1743-422X-10-348. Virol J. 2013. PMID: 24295514 Free PMC article.
-
Direct antigen presentation is the canonical pathway of cytomegalovirus CD8 T-cell priming regulated by balanced immune evasion ensuring a strong antiviral response.Front Immunol. 2023 Dec 12;14:1272166. doi: 10.3389/fimmu.2023.1272166. eCollection 2023. Front Immunol. 2023. PMID: 38149242 Free PMC article.
-
The Canonical Immediate Early 3 Gene Product pIE611 of Mouse Cytomegalovirus Is Dispensable for Viral Replication but Mediates Transcriptional and Posttranscriptional Regulation of Viral Gene Products.J Virol. 2015 Aug;89(16):8590-8. doi: 10.1128/JVI.01234-15. Epub 2015 Jun 10. J Virol. 2015. PMID: 26063418 Free PMC article.
-
Prolonged activation of cytomegalovirus early gene e1-promoter exclusively in neurons during infection of the developing cerebrum.Acta Neuropathol Commun. 2021 Mar 9;9(1):39. doi: 10.1186/s40478-021-01139-0. Acta Neuropathol Commun. 2021. PMID: 33750455 Free PMC article.
References
-
- Mocarski ES, Jr, Shenk T, Pass RF. 2006. Cytomegaloviruses, 5th ed Lippincott Williams &Wilkins, Philadelphia, PA
-
- Reddehase MJ, Simon CO, Seckert CK, Lemmermann N, Grzimek NK. 2008. Murine model of cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325:315–331 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

