Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation
- PMID: 23238393
- PMCID: PMC3596133
- DOI: 10.1038/embor.2012.208
Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation
Abstract
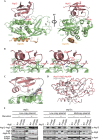
Atg12 is conjugated to Atg5 through enzymatic reactions similar to ubiquitination. The Atg12-Atg5 conjugate functions as an E3-like enzyme to promote lipidation of Atg8, whereas lipidated Atg8 has essential roles in both autophagosome formation and selective cargo recognition during autophagy. However, the molecular role of Atg12 modification in these processes has remained elusive. Here, we report the crystal structure of the Atg12-Atg5 conjugate. In addition to the isopeptide linkage, Atg12 forms hydrophobic and hydrophilic interactions with Atg5, thereby fixing its position on Atg5. Structural comparison with unmodified Atg5 and mutational analyses showed that Atg12 modification neither induces a conformational change in Atg5 nor creates a functionally important architecture. Rather, Atg12 functions as a binding module for Atg3, the E2 enzyme for Atg8, thus endowing Atg5 with the ability to interact with Atg3 to facilitate Atg8 lipidation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy.J Biol Chem. 2007 Dec 28;282(52):37298-302. doi: 10.1074/jbc.C700195200. Epub 2007 Nov 6. J Biol Chem. 2007. PMID: 17986448
-
Atg12-Interacting Motif Is Crucial for E2-E3 Interaction in Plant Atg8 System.Biol Pharm Bull. 2021 Sep 1;44(9):1337-1343. doi: 10.1248/bpb.b21-00439. Epub 2021 Jul 1. Biol Pharm Bull. 2021. PMID: 34193767
-
Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation.Autophagy. 2013 Mar;9(3):424-5. doi: 10.4161/auto.22931. Epub 2013 Jan 15. Autophagy. 2013. PMID: 23321721 Free PMC article.
-
The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series.EMBO Rep. 2008 Sep;9(9):859-64. doi: 10.1038/embor.2008.163. EMBO Rep. 2008. PMID: 18704115 Free PMC article. Review.
-
Beyond Atg8 binding: The role of AIM/LIR motifs in autophagy.Autophagy. 2017 May 4;13(5):978-979. doi: 10.1080/15548627.2016.1277311. Epub 2017 Jan 25. Autophagy. 2017. PMID: 28121222 Free PMC article. Review.
Cited by
-
The IRE1α pathway in glomerular diseases: The unfolded protein response and beyond.Front Mol Med. 2022 Sep 26;2:971247. doi: 10.3389/fmmed.2022.971247. eCollection 2022. Front Mol Med. 2022. PMID: 39086958 Free PMC article. Review.
-
Dihydroorotase MoPyr4 is required for development, pathogenicity, and autophagy in rice blast fungus.Cell Commun Signal. 2024 Jul 15;22(1):362. doi: 10.1186/s12964-024-01741-4. Cell Commun Signal. 2024. PMID: 39010102 Free PMC article.
-
When an underdog becomes a major player: the role of protein structural disorder in the Atg8 conjugation system.Autophagy. 2024 Oct;20(10):2338-2345. doi: 10.1080/15548627.2024.2357496. Epub 2024 May 29. Autophagy. 2024. PMID: 38808635 Review.
-
Mutations accumulated in the Spike of SARS-CoV-2 Omicron allow for more efficient counteraction of the restriction factor BST2/Tetherin.PLoS Pathog. 2024 Jan 8;20(1):e1011912. doi: 10.1371/journal.ppat.1011912. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38190411 Free PMC article.
-
ATG8f Interacts with Chilli Veinal Mottle Virus 6K2 Protein to Limit Virus Infection.Viruses. 2023 Nov 26;15(12):2324. doi: 10.3390/v15122324. Viruses. 2023. PMID: 38140565 Free PMC article.
References
-
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 - PubMed
-
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y (1998) A protein conjugation system essential for autophagy. Nature 395: 395–398 - PubMed
-
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132 - PubMed
-
- Noda NN, Ohsumi Y, Inagaki F (2009) ATG systems from the protein structural point of view. Chem Rev 109: 1587–1598 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

