Identification of interdependent variables that influence coreceptor switch in R5 SHIV(SF162P3N)-infected macaques
- PMID: 23237529
- PMCID: PMC3528637
- DOI: 10.1186/1742-4690-9-106
Identification of interdependent variables that influence coreceptor switch in R5 SHIV(SF162P3N)-infected macaques
Abstract
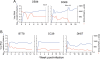
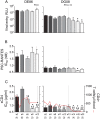
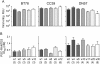
Background: We previously reported that adoption of an "open" envelope glycoprotein (Env) to expose the CD4 binding site for efficient receptor binding and infection of cell targets such as macrophages that express low levels of the receptor represents an early event in the process of coreceptor switch in two rapidly progressing (RP) R5 SHIV(SF162P3N)-infected rhesus macaques, releasing or reducing Env structural constraints that have been suggested to limit the pathways available for a change in coreceptor preference. Here we extended these studies to two additional RP monkeys with coreceptor switch and three without to confirm and identify additional factors that facilitated the process of phenotypic conversion.
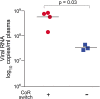
Results: We found that regardless of coreceptor switching, R5 viruses in SHIV(SF162P3N)-infected RP macaques evolved over time to infect macrophages more efficiently; this was accompanied by increased sCD4 sensitivity, with structural changes in the CD4 binding site, the V3 loop and/or the fusion domain of their Envs that are suggestive of better CD4 contact, CCR5 usage and/or virus fusion. However, sCD4-sensitive variants with improved CD4 binding were observed only in RPs with coreceptor switch. Furthermore, cumulative viral load was higher in RPs with than in those without phenotypic switch, with the latter maintaining a longer period of seroconversion.
Conclusions: Our data suggest that the increased virus replication in the RPs with R5-to-X4 conversion increased the rate of virus evolution and reduction in the availability of target cells with optimal CD4 expression heightened the competition for binding to the receptor. In the absence of immunological restrictions, variants that adopt an "open" Env to expose the CD4 binding site for better CD4 use are selected, allowing structural changes that confer CXCR4-use to be manifested. Viral load, change in target cell population during the course of infection and host immune response therefore are interdependent variables that influence R5 virus evolution and coreceptor switch in SHIV(SF162P3N)-infected rhesus macaques. Because an "open" Env conformation also renders the virus more susceptible to antibody neutralization, our findings help to explain the infrequent and late appearance of X4 virus in HIV-1 infection when the immune system deteriorates.
Figures
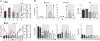

Similar articles
-
Adoption of an "open" envelope conformation facilitating CD4 binding and structural remodeling precedes coreceptor switch in R5 SHIV-infected macaques.PLoS One. 2011;6(7):e21350. doi: 10.1371/journal.pone.0021350. Epub 2011 Jul 8. PLoS One. 2011. PMID: 21760891 Free PMC article.
-
V3 loop-determined coreceptor preference dictates the dynamics of CD4+-T-cell loss in simian-human immunodeficiency virus-infected macaques.J Virol. 2005 Oct;79(19):12296-303. doi: 10.1128/JVI.79.19.12296-12303.2005. J Virol. 2005. PMID: 16160156 Free PMC article.
-
R5X4 viruses are evolutionary, functional, and antigenic intermediates in the pathway of a simian-human immunodeficiency virus coreceptor switch.J Virol. 2008 Jul;82(14):7089-99. doi: 10.1128/JVI.00570-08. Epub 2008 May 14. J Virol. 2008. PMID: 18480460 Free PMC article.
-
Coreceptor switch in infection of nonhuman primates.Curr HIV Res. 2009 Jan;7(1):30-8. doi: 10.2174/157016209787048500. Curr HIV Res. 2009. PMID: 19149552 Review.
-
The SIV Envelope Glycoprotein, Viral Tropism, and Pathogenesis: Novel Insights from Nonhuman Primate Models of AIDS.Curr HIV Res. 2018;16(1):29-40. doi: 10.2174/1570162X15666171124123116. Curr HIV Res. 2018. PMID: 29173176 Review.
Cited by
-
High-Sequence Diversity and Rapid Virus Turnover Contribute to Higher Rates of Coreceptor Switching in Treatment-Experienced Subjects with HIV-1 Viremia.AIDS Res Hum Retroviruses. 2017 Mar;33(3):234-245. doi: 10.1089/AID.2016.0153. Epub 2016 Oct 12. AIDS Res Hum Retroviruses. 2017. PMID: 27604829 Free PMC article.
-
Existence of Replication-Competent Minor Variants with Different Coreceptor Usage in Plasma from HIV-1-Infected Individuals.J Virol. 2020 Jun 1;94(12):e00193-20. doi: 10.1128/JVI.00193-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32295903 Free PMC article.
-
Stepping toward a macaque model of HIV-1 induced AIDS.Viruses. 2014 Sep 25;6(9):3643-51. doi: 10.3390/v6093643. Viruses. 2014. PMID: 25256394 Free PMC article. Review.
-
Antigenic and 3D structural characterization of soluble X4 and hybrid X4-R5 HIV-1 Env trimers.Retrovirology. 2014 May 30;11:42. doi: 10.1186/1742-4690-11-42. Retrovirology. 2014. PMID: 24884925 Free PMC article.
-
The selection of low envelope glycoprotein reactivity to soluble CD4 and cold during simian-human immunodeficiency virus infection of rhesus macaques.J Virol. 2014 Jan;88(1):21-40. doi: 10.1128/JVI.01558-13. Epub 2013 Oct 16. J Virol. 2014. PMID: 24131720 Free PMC article.
References
-
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H. et al.Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. - DOI - PMC - PubMed
-
- Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH. et al.Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. - DOI - PMC - PubMed
-
- Koot M, Keet IP, Vos AH, de Goede RE, Roos MT, Coutinho RA, Miedema F, Schellekens PT, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

