Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids
- PMID: 23230278
- PMCID: PMC3591653
- DOI: 10.1074/mcp.M112.021303
Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids
Abstract
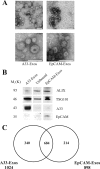
Exosomes are naturally occurring biological nanomembranous vesicles (∼40 to 100 nm) of endocytic origin that are released from diverse cell types into the extracellular space. They have pleiotropic functions such as antigen presentation and intercellular transfer of protein cargo, mRNA, microRNA, lipids, and oncogenic potential. Here we describe the isolation, via sequential immunocapture using anti-A33- and anti-EpCAM-coupled magnetic beads, of two distinct populations of exosomes released from organoids derived from human colon carcinoma cell line LIM1863. The exosome populations (A33-Exos and EpCAM-Exos) could not be distinguished via electron microscopy and contained stereotypical exosome markers such as TSG101, Alix, and HSP70. The salient finding of this study, revealed via gel-based LC-MS/MS, was the exclusive identification in EpCAM-Exos of the classical apical trafficking molecules CD63 (LAMP3), mucin 13 and the apical intestinal enzyme sucrase isomaltase and increased expression of dipeptidyl peptidase IV and the apically restricted pentaspan membrane glycoprotein prominin 1. In contrast, the A33-Exos preparation was enriched with basolateral trafficking molecules such as early endosome antigen 1, the Golgi membrane protein ADP-ribosylation factor, and clathrin. Our observations are consistent with EpCAM- and A33-Exos being released from the apical and basolateral surfaces, respectively, and the EpCAM-Exos proteome profile with widely published stereotypical exosomes. A proteome analysis of LIM1863-derived shed microvesicles (sMVs) was also performed in order to clearly distinguish A33- and EpCAM-Exos from sMVs. Intriguingly, several members of the MHC class I family of antigen presentation molecules were exclusively observed in A33-Exos, whereas neither MHC class I nor MHC class II molecules were observed via MS in EpCAM-Exos. Additionally, we report for the first time in any extracellular vesicle study the colocalization of EpCAM, claudin-7, and CD44 in EpCAM-Exos. Given that these molecules are known to complex together to promote tumor progression, further characterization of exosome subpopulations will enable a deeper understanding of their possible role in regulation of the tumor microenvironment.
Figures


Similar articles
-
Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct.Methods. 2015 Oct 1;87:11-25. doi: 10.1016/j.ymeth.2015.04.008. Epub 2015 Apr 16. Methods. 2015. PMID: 25890246
-
Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes.Methods. 2012 Feb;56(2):293-304. doi: 10.1016/j.ymeth.2012.01.002. Epub 2012 Jan 21. Methods. 2012. PMID: 22285593
-
Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature.Mol Cell Proteomics. 2010 Feb;9(2):197-208. doi: 10.1074/mcp.M900152-MCP200. Epub 2009 Oct 16. Mol Cell Proteomics. 2010. PMID: 19837982 Free PMC article.
-
CD44 and EpCAM: cancer-initiating cell markers.Curr Mol Med. 2008 Dec;8(8):784-804. doi: 10.2174/156652408786733667. Curr Mol Med. 2008. PMID: 19075676 Review.
-
The tumor antigen EpCAM: tetraspanins and the tight junction protein claudin-7, new partners, new functions.Front Biosci. 2008 May 1;13:5847-65. doi: 10.2741/3121. Front Biosci. 2008. PMID: 18508627 Review.
Cited by
-
Information Transfer and Biological Significance of Neoplastic Exosomes in the Tumor Microenvironment of Osteosarcoma.Onco Targets Ther. 2020 Sep 8;13:8931-8940. doi: 10.2147/OTT.S266835. eCollection 2020. Onco Targets Ther. 2020. PMID: 32982285 Free PMC article. Review.
-
Exosomes mediate cell contact-independent ephrin-Eph signaling during axon guidance.J Cell Biol. 2016 Jul 4;214(1):35-44. doi: 10.1083/jcb.201601085. Epub 2016 Jun 27. J Cell Biol. 2016. PMID: 27354374 Free PMC article.
-
Extracellular vesicles and particles impact the systemic landscape of cancer.EMBO J. 2022 Sep 15;41(18):e109288. doi: 10.15252/embj.2021109288. Epub 2022 Sep 2. EMBO J. 2022. PMID: 36052513 Free PMC article. Review.
-
Biochemical and biological characterization of exosomes containing prominin-1/CD133.Mol Cancer. 2013 Jun 14;12:62. doi: 10.1186/1476-4598-12-62. Mol Cancer. 2013. PMID: 23767874 Free PMC article.
-
Visualizing Extracellular Vesicles and Their Function in 3D Tumor Microenvironment Models.Int J Mol Sci. 2021 Apr 30;22(9):4784. doi: 10.3390/ijms22094784. Int J Mol Sci. 2021. PMID: 33946403 Free PMC article. Review.
References
-
- Mueller M. M., Fusenig N. E. (2004) Friends or foes—bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer 4, 839–849 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

