Comparison of genome-wide binding of MyoD in normal human myogenic cells and rhabdomyosarcomas identifies regional and local suppression of promyogenic transcription factors
- PMID: 23230269
- PMCID: PMC3571334
- DOI: 10.1128/MCB.00916-12
Comparison of genome-wide binding of MyoD in normal human myogenic cells and rhabdomyosarcomas identifies regional and local suppression of promyogenic transcription factors
Abstract
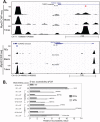
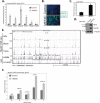
Rhabdomyosarcoma is a pediatric tumor of skeletal muscle that expresses the myogenic basic helix-loop-helix protein MyoD but fails to undergo terminal differentiation. Prior work has determined that DNA binding by MyoD occurs in the tumor cells, but myogenic targets fail to activate. Using MyoD chromatin immunoprecipitation coupled to high-throughput sequencing and gene expression analysis in both primary human muscle cells and RD rhabdomyosarcoma cells, we demonstrate that MyoD binds in a similar genome-wide pattern in both tumor and normal cells but binds poorly at a subset of myogenic genes that fail to activate in the tumor cells. Binding differences are found both across genomic regions and locally at specific sites that are associated with binding motifs for RUNX1, MEF2C, JDP2, and NFIC. These factors are expressed at lower levels in RD cells than muscle cells and rescue myogenesis when expressed in RD cells. MEF2C is located in a genomic region that exhibits poor MyoD binding in RD cells, whereas JDP2 exhibits local DNA hypermethylation in its promoter in both RD cells and primary tumor samples. These results demonstrate that regional and local silencing of differentiation factors contributes to the differentiation defect in rhabdomyosarcomas.
Figures




Similar articles
-
The ablation of EZH2 uncovers its crucial role in rhabdomyosarcoma formation.Cell Cycle. 2012 Oct 15;11(20):3828-36. doi: 10.4161/cc.22025. Epub 2012 Sep 14. Cell Cycle. 2012. PMID: 22983009 Free PMC article.
-
Twist2 amplification in rhabdomyosarcoma represses myogenesis and promotes oncogenesis by redirecting MyoD DNA binding.Genes Dev. 2019 Jun 1;33(11-12):626-640. doi: 10.1101/gad.324467.119. Epub 2019 Apr 11. Genes Dev. 2019. PMID: 30975722 Free PMC article.
-
The transcriptional co-repressor TLE3 regulates myogenic differentiation by repressing the activity of the MyoD transcription factor.J Biol Chem. 2017 Aug 4;292(31):12885-12894. doi: 10.1074/jbc.M116.774570. Epub 2017 Jun 12. J Biol Chem. 2017. PMID: 28607151 Free PMC article.
-
[New insight into MyoD regulation: involvement in rhabdomyosarcoma pathway?].Bull Cancer. 2001 Jun;88(6):545-8. Bull Cancer. 2001. PMID: 11459700 Review. French.
-
MicroRNAs in rhabdomyosarcoma: pathogenetic implications and translational potentiality.Mol Cancer. 2011 Sep 24;10:120. doi: 10.1186/1476-4598-10-120. Mol Cancer. 2011. PMID: 21943149 Free PMC article. Review.
Cited by
-
Targeting DNA Methylation Machinery in Pediatric Solid Tumors.Cells. 2024 Jul 18;13(14):1209. doi: 10.3390/cells13141209. Cells. 2024. PMID: 39056791 Free PMC article. Review.
-
A dysfunctional miR-1-TRPS1-MYOG axis drives ERMS by suppressing terminal myogenic differentiation.Mol Ther. 2023 Sep 6;31(9):2612-2632. doi: 10.1016/j.ymthe.2023.07.003. Epub 2023 Jul 14. Mol Ther. 2023. PMID: 37452493 Free PMC article.
-
TWIST2-mediated chromatin remodeling promotes fusion-negative rhabdomyosarcoma.Sci Adv. 2023 Apr 28;9(17):eade8184. doi: 10.1126/sciadv.ade8184. Epub 2023 Apr 28. Sci Adv. 2023. PMID: 37115930 Free PMC article.
-
Pediatric cancer epigenome and the influence of folate.Epigenomics. 2015;7(6):961-73. doi: 10.2217/epi.15.42. Epub 2015 May 7. Epigenomics. 2015. PMID: 25950259 Free PMC article. Review.
-
CRISPR screen identifies the NCOR/HDAC3 complex as a major suppressor of differentiation in rhabdomyosarcoma.Proc Natl Acad Sci U S A. 2016 Dec 27;113(52):15090-15095. doi: 10.1073/pnas.1610270114. Epub 2016 Dec 12. Proc Natl Acad Sci U S A. 2016. PMID: 27956629 Free PMC article.
References
-
- Lassar AB, Buskin JN, Lockshon D, Davis RL, Apone S, Hauschka SD, Weintraub H. 1989. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell 58:823–831 - PubMed
-
- Murre C, McCaw PS, Baltimore D. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777–783 - PubMed
-
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66:305–315 - PubMed
-
- Merlino G, Helman L. 1999. Rhabdomyosarcoma—working out the pathways. Oncogene 18:5340–5348 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
