A uvs-5 strain is deficient for a mitofusin gene homologue, fzo1, involved in maintenance of long life span in Neurospora crassa
- PMID: 23223037
- PMCID: PMC3571291
- DOI: 10.1128/EC.00226-12
A uvs-5 strain is deficient for a mitofusin gene homologue, fzo1, involved in maintenance of long life span in Neurospora crassa
Abstract
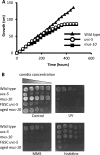
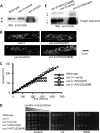
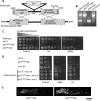
Mitochondria are highly dynamic organelles that continuously fuse and divide. To maintain mitochondria, cells establish an equilibrium of fusion and fission events, which are mediated by dynamin-like GTPases. We previously showed that an mus-10 strain, a mutagen-sensitive strain of the filamentous fungus Neurospora crassa, is defective in an F-box protein that is essential for the maintenance of mitochondrial DNA (mtDNA), long life span, and mitochondrial morphology. Similarly, a uvs-5 mutant accumulates deletions within its mtDNA, has a shortened life span, and harbors fragmented mitochondria, the latter of which is indicative of an imbalance between mitochondrial fission and fusion. Since the uvs-5 mutation maps very close to the locus of fzo1, encoding a mitofusin homologue thought to mediate mitochondrial outer membrane fusion, we determined the sequence of the fzo1 gene in the uvs-5 mutant. A single amino acid substitution (Q368R) was found in the GTPase domain of the FZO1 protein. Expression of wild-type FZO1 in the uvs-5 strain rescued the mutant phenotypes, while expression of a mutant FZO1 protein did not. Moreover, when knock-in of the Q368R mutation was performed on a wild-type strain, the resulting mutant displayed phenotypes identical to those of the uvs-5 mutant. Therefore, we concluded that the previously unidentified uvs-5 gene is fzo1. Furthermore, we used immunoprecipitation analysis to show that the FZO1 protein interacts with MUS-10, which suggests that these two proteins may function together to maintain mitochondrial morphology.
Figures



Similar articles
-
Sequential requirements for the GTPase domain of the mitofusin Fzo1 and the ubiquitin ligase SCFMdm30 in mitochondrial outer membrane fusion.J Cell Sci. 2011 May 1;124(Pt 9):1403-10. doi: 10.1242/jcs.079293. J Cell Sci. 2011. PMID: 21502136 Free PMC article.
-
Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae.J Biol Chem. 2005 May 13;280(19):18598-603. doi: 10.1074/jbc.M500807200. Epub 2005 Mar 10. J Biol Chem. 2005. PMID: 15760898
-
A mutation associated with CMT2A neuropathy causes defects in Fzo1 GTP hydrolysis, ubiquitylation, and protein turnover.Mol Biol Cell. 2009 Dec;20(23):5026-35. doi: 10.1091/mbc.e09-07-0622. Epub 2009 Oct 7. Mol Biol Cell. 2009. PMID: 19812251 Free PMC article.
-
Ugo1 and Mdm30 act sequentially during Fzo1-mediated mitochondrial outer membrane fusion.J Cell Sci. 2011 Apr 1;124(Pt 7):1126-35. doi: 10.1242/jcs.073080. Epub 2011 Mar 8. J Cell Sci. 2011. PMID: 21385840
-
The Neurospora crassa UVS-3 epistasis group encodes homologues of the ATR/ATRIP checkpoint control system.DNA Repair (Amst). 2008 Feb 1;7(2):213-29. doi: 10.1016/j.dnarep.2007.09.011. Epub 2007 Nov 5. DNA Repair (Amst). 2008. PMID: 17983847
Cited by
-
Neural activity and CaMKII protect mitochondria from fragmentation in aging Caenorhabditis elegans neurons.Proc Natl Acad Sci U S A. 2015 Jul 14;112(28):8768-73. doi: 10.1073/pnas.1501831112. Epub 2015 Jun 29. Proc Natl Acad Sci U S A. 2015. PMID: 26124107 Free PMC article.
-
Inheritance of the fittest mitochondria in yeast.Trends Cell Biol. 2014 Jan;24(1):53-60. doi: 10.1016/j.tcb.2013.07.003. Epub 2013 Aug 6. Trends Cell Biol. 2014. PMID: 23932848 Free PMC article. Review.
-
Quercetin-Induced Lifespan Extension in Podospora anserina Requires Methylation of the Flavonoid by the O-Methyltransferase PaMTH1.Front Genet. 2018 May 4;9:160. doi: 10.3389/fgene.2018.00160. eCollection 2018. Front Genet. 2018. PMID: 29780405 Free PMC article.
References
-
- Osiewacz HD. 2002. Genes, mitochondria and aging in filamentous fungi. Ageing Res. Rev. 1:425–442 - PubMed
-
- Lorin S, Dufour E, Boulay J, Begel O, Marsy S, Sainsard-Chanet A. 2001. Overexpression of the alternative oxidase restores senescence and fertility in a long-lived respiration-deficient mutant of Podospora anserina. Mol. Microbiol. 42:1259–1267 - PubMed
-
- Espagne E, Lespinet O, Malagnac F, Da Silva C, Jaillon O, Porcel BM, Couloux A, Aury JM, Segurens B, Poulain J, Anthouard V, Grossetete S, Khalili H, Coppin E, Dequard-Chablat M, Picard M, Contamine V, Arnaise S, Bourdais A, Berteaux-Lecellier V, Gautheret D, de Vries RP, Battaglia E, Coutinho PM, Danchin EG, Henrissat B, Khoury RE, Sainsard-Chanet A, Boivin A, Pinan-Lucarre B, Sellem CH, Debuchy R, Wincker P, Weissenbach J, Silar P. 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9:R77 doi:10.1186/gb-2008-9-5-r77 - DOI - PMC - PubMed
-
- Maheshwari R, Navaraj A. 2008. Senescence in fungi: the view from Neurospora. FEMS Microbiol. Lett. 280:135–143 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

