Activity of Proteus mirabilis FliL is viscosity dependent and requires extragenic DNA
- PMID: 23222728
- PMCID: PMC3562113
- DOI: 10.1128/JB.02024-12
Activity of Proteus mirabilis FliL is viscosity dependent and requires extragenic DNA
Abstract
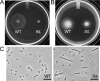
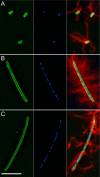
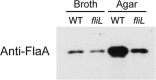
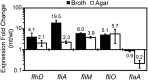
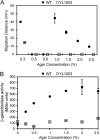
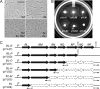
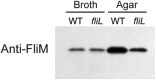
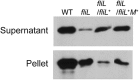
Proteus mirabilis is a urinary tract pathogen and well known for its ability to move over agar surfaces by flagellum-dependent swarming motility. When P. mirabilis encounters a highly viscous environment, e.g., an agar surface, it differentiates from short rods with few flagella to elongated, highly flagellated cells that lack septa and contain multiple nucleoids. The bacteria detect a surface by monitoring the rotation of their flagellar motors. This process involves an enigmatic flagellar protein called FliL, the first gene in an operon (fliLMNOPQR) that encodes proteins of the flagellar rotor switch complex and flagellar export apparatus. We used a fliL knockout mutant to gain further insight into the function of FliL. Loss of FliL results in cells that cannot swarm (Swr(-)) but do swim (Swm(+)) and produces cells that look like wild-type swarmer cells, termed "pseudoswarmer cells," that are elongated, contain multiple nucleoids, and lack septa. Unlike swarmer cells, pseudoswarmer cells are not hyperflagellated due to reduced expression of flaA (the gene encoding flagellin), despite an increased transcription of both flhD and fliA, two positive regulators of flagellar gene expression. We found that defects in fliL prevent viscosity-dependent sensing of a surface and viscosity-dependent induction of flaA transcription. Studies with fliL cells unexpectedly revealed that the fliL promoter, fliL coding region, and a portion of fliM DNA are needed to complement the Swr(-) phenotype. The data support a dual role for FliL as a critical link in sensing a surface and in the maintenance of flagellar rod integrity.
Figures
Similar articles
-
Loss of FliL alters Proteus mirabilis surface sensing and temperature-dependent swarming.J Bacteriol. 2015 Jan 1;197(1):159-73. doi: 10.1128/JB.02235-14. Epub 2014 Oct 20. J Bacteriol. 2015. PMID: 25331431 Free PMC article.
-
Perturbation of FliL interferes with Proteus mirabilis swarmer cell gene expression and differentiation.J Bacteriol. 2012 Jan;194(2):437-47. doi: 10.1128/JB.05998-11. Epub 2011 Nov 11. J Bacteriol. 2012. PMID: 22081397 Free PMC article.
-
The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein.J Bacteriol. 2005 Oct;187(19):6789-803. doi: 10.1128/JB.187.19.6789-6803.2005. J Bacteriol. 2005. PMID: 16166542 Free PMC article.
-
Regulation of gene expression during swarmer cell differentiation in Proteus mirabilis.FEMS Microbiol Rev. 2010 Sep;34(5):753-63. doi: 10.1111/j.1574-6976.2010.00229.x. Epub 2010 Apr 19. FEMS Microbiol Rev. 2010. PMID: 20497230 Review.
-
Flagellar protein FliL: A many-splendored thing.Mol Microbiol. 2024 Oct;122(4):447-454. doi: 10.1111/mmi.15301. Epub 2024 Aug 3. Mol Microbiol. 2024. PMID: 39096095 Free PMC article. Review.
Cited by
-
A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis.Mol Microbiol. 2013 Oct;90(1):6-21. doi: 10.1111/mmi.12342. Epub 2013 Aug 14. Mol Microbiol. 2013. PMID: 23888912 Free PMC article.
-
A distant homologue of the FlgT protein interacts with MotB and FliL and is essential for flagellar rotation in Rhodobacter sphaeroides.J Bacteriol. 2013 Dec;195(23):5285-96. doi: 10.1128/JB.00760-13. Epub 2013 Sep 20. J Bacteriol. 2013. PMID: 24056105 Free PMC article.
-
Proteus mirabilis and Urinary Tract Infections.Microbiol Spectr. 2015 Oct;3(5):10.1128/microbiolspec.UTI-0017-2013. doi: 10.1128/microbiolspec.UTI-0017-2013. Microbiol Spectr. 2015. PMID: 26542036 Free PMC article. Review.
-
Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis.J Bacteriol. 2014 Feb;196(4):740-53. doi: 10.1128/JB.01217-13. Epub 2013 Dec 2. J Bacteriol. 2014. PMID: 24296669 Free PMC article.
-
Functional Regulators of Bacterial Flagella.Annu Rev Microbiol. 2019 Sep 8;73:225-246. doi: 10.1146/annurev-micro-020518-115725. Epub 2019 May 28. Annu Rev Microbiol. 2019. PMID: 31136265 Free PMC article. Review.
References
-
- Mobley HL, Belas R. 1995. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 3:280–284 - PubMed
-
- Alavi M, Belas R. 2001. Surface sensing, swarmer cell differentiation, and biofilm development. Methods Enzymol. 336:29–40 - PubMed
-
- Allison C, Lai HC, Hughes C. 1992. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6:1583–1591 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

