Promoter cross-talk via a shared enhancer explains paternally biased expression of Nctc1 at the Igf2/H19/Nctc1 imprinted locus
- PMID: 23221643
- PMCID: PMC3553941
- DOI: 10.1093/nar/gks1182
Promoter cross-talk via a shared enhancer explains paternally biased expression of Nctc1 at the Igf2/H19/Nctc1 imprinted locus
Abstract
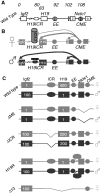
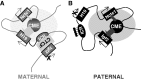
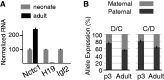
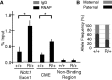
Developmentally regulated transcription often depends on physical interactions between distal enhancers and their cognate promoters. Recent genomic analyses suggest that promoter-promoter interactions might play a similarly critical role in organizing the genome and establishing cell-type-specific gene expression. The Igf2/H19 locus has been a valuable model for clarifying the role of long-range interactions between cis-regulatory elements. Imprinted expression of the linked, reciprocally imprinted genes is explained by parent-of-origin-specific chromosomal loop structures between the paternal Igf2 or maternal H19 promoters and their shared tissue-specific enhancer elements. Here, we further analyze these loop structures for their composition and their impact on expression of the linked long non-coding RNA, Nctc1. We show that Nctc1 is co-regulated with Igf2 and H19 and physically interacts with the shared muscle enhancer. In fact, all three co-regulated genes have the potential to interact not only with the shared enhancer but also with each other via their enhancer interactions. Furthermore, developmental and genetic analyses indicate functional significance for these promoter-promoter interactions. Altogether, we present a novel mechanism to explain developmental specific imprinting of Nctc1 and provide new information about enhancer mechanisms and about the role of chromatin domains in establishing gene expression patterns.
Figures
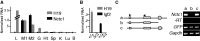
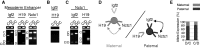
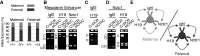
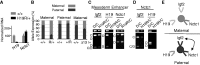
Similar articles
-
Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking.Hum Mol Genet. 2008 Oct 1;17(19):3021-9. doi: 10.1093/hmg/ddn200. Epub 2008 Jul 10. Hum Mol Genet. 2008. PMID: 18617529 Free PMC article.
-
Regulatory mechanisms at the mouse Igf2/H19 locus.Mol Cell Biol. 2001 Dec;21(23):8189-96. doi: 10.1128/MCB.21.23.8189-8196.2001. Mol Cell Biol. 2001. PMID: 11689707 Free PMC article.
-
Long-range chromatin interactions at the mouse Igf2/H19 locus reveal a novel paternally expressed long non-coding RNA.Nucleic Acids Res. 2011 Aug;39(14):5893-906. doi: 10.1093/nar/gkr209. Epub 2011 Apr 7. Nucleic Acids Res. 2011. PMID: 21478171 Free PMC article.
-
Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation.J Biochem. 2000 May;127(5):711-5. doi: 10.1093/oxfordjournals.jbchem.a022661. J Biochem. 2000. PMID: 10788777 Review.
-
H19 and Igf2--enhancing the confusion?Trends Genet. 2003 Jan;19(1):17-23. doi: 10.1016/s0168-9525(02)00004-5. Trends Genet. 2003. PMID: 12493244 Review.
Cited by
-
The complex genetics of human insulin-like growth factor 2 are not reflected in public databases.J Biol Chem. 2018 Mar 23;293(12):4324-4333. doi: 10.1074/jbc.RA117.001573. Epub 2018 Feb 2. J Biol Chem. 2018. PMID: 29414792 Free PMC article.
-
Exploring chromatin structural roles of non-coding RNAs at imprinted domains.Biochem Soc Trans. 2021 Aug 27;49(4):1867-1879. doi: 10.1042/BST20210758. Biochem Soc Trans. 2021. PMID: 34338292 Free PMC article. Review.
-
Differential 3D genome architecture and imprinted gene expression: cause or consequence?Biochem Soc Trans. 2024 Jun 26;52(3):973-986. doi: 10.1042/BST20230143. Biochem Soc Trans. 2024. PMID: 38775198 Free PMC article. Review.
-
H19ICR mediated transcriptional silencing does not require target promoter methylation.Biochem Biophys Res Commun. 2016 Jul 29;476(3):121-6. doi: 10.1016/j.bbrc.2016.05.042. Epub 2016 May 10. Biochem Biophys Res Commun. 2016. PMID: 27178213 Free PMC article.
-
Human IGF2 Gene Epigenetic and Transcriptional Regulation: At the Core of Developmental Growth and Tumorigenic Behavior.Biomedicines. 2023 Jun 7;11(6):1655. doi: 10.3390/biomedicines11061655. Biomedicines. 2023. PMID: 37371750 Free PMC article. Review.
References
-
- Ainscough JF-X, Dandola L, Surani MA. Appropriate expression of the mouse H19 gene utilises three or more distinct enhancer regions spread over more than 130 kb. Mech. Dev. 2000;91:365–368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

