Protein secondary structure determination by constrained single-particle cryo-electron tomography
- PMID: 23217682
- PMCID: PMC3600145
- DOI: 10.1016/j.str.2012.10.016
Protein secondary structure determination by constrained single-particle cryo-electron tomography
Abstract
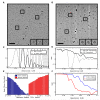
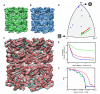
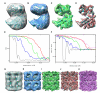
Cryo-electron microscopy (cryo-EM) is a powerful technique for 3D structure determination of protein complexes by averaging information from individual molecular images. The resolutions that can be achieved with single-particle cryo-EM are frequently limited by inaccuracies in assigning molecular orientations based solely on 2D projection images. Tomographic data collection schemes, however, provide powerful constraints that can be used to more accurately determine molecular orientations necessary for 3D reconstruction. Here, we propose "constrained single-particle tomography" as a general strategy for 3D structure determination in cryo-EM. A key component of our approach is the effective use of images recorded in tilt series to extract high-resolution information and correct for the contrast transfer function. By incorporating geometric constraints into the refinement to improve orientational accuracy of images, we reduce model bias and overrefinement artifacts and demonstrate that protein structures can be determined at resolutions of ∼8 Å starting from low-dose tomographic tilt series.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
IPET and FETR: experimental approach for studying molecular structure dynamics by cryo-electron tomography of a single-molecule structure.PLoS One. 2012;7(1):e30249. doi: 10.1371/journal.pone.0030249. Epub 2012 Jan 24. PLoS One. 2012. PMID: 22291925 Free PMC article.
-
Cryo-Electron Tomography and Subtomogram Averaging.Methods Enzymol. 2016;579:329-67. doi: 10.1016/bs.mie.2016.04.014. Epub 2016 Jun 22. Methods Enzymol. 2016. PMID: 27572733 Review.
-
High-resolution single-particle 3D analysis on GroEL prepared by cryo-negative staining.Micron. 2008 Oct;39(7):934-43. doi: 10.1016/j.micron.2007.11.003. Epub 2007 Nov 17. Micron. 2008. PMID: 18083582
-
Cryo-electron microscopy and cryo-electron tomography of nanoparticles.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2017 Mar;9(2). doi: 10.1002/wnan.1417. Epub 2016 Jun 23. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2017. PMID: 27339510 Review.
-
emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging.Nat Methods. 2018 Nov;15(11):955-961. doi: 10.1038/s41592-018-0167-z. Epub 2018 Oct 22. Nat Methods. 2018. PMID: 30349041 Free PMC article.
Cited by
-
A New Protocol for Atomic-Level Protein Structure Modeling and Refinement Using Low-to-Medium Resolution Cryo-EM Density Maps.J Mol Biol. 2020 Sep 4;432(19):5365-5377. doi: 10.1016/j.jmb.2020.07.027. Epub 2020 Aug 6. J Mol Biol. 2020. PMID: 32771523 Free PMC article.
-
CryoDRGN-ET: deep reconstructing generative networks for visualizing dynamic biomolecules inside cells.Nat Methods. 2024 Aug;21(8):1537-1545. doi: 10.1038/s41592-024-02340-4. Epub 2024 Jul 18. Nat Methods. 2024. PMID: 39025970
-
Cryo-electron microscopy--a primer for the non-microscopist.FEBS J. 2013 Jan;280(1):28-45. doi: 10.1111/febs.12078. Epub 2012 Dec 17. FEBS J. 2013. PMID: 23181775 Free PMC article. Review.
-
Subnanometer-resolution structure determination in situ by hybrid subtomogram averaging - single particle cryo-EM.Nat Commun. 2020 Jul 24;11(1):3709. doi: 10.1038/s41467-020-17466-0. Nat Commun. 2020. PMID: 32709843 Free PMC article.
-
High-resolution cryo-electron microscopy on macromolecular complexes and cell organelles.Protoplasma. 2014 Mar;251(2):417-27. doi: 10.1007/s00709-013-0600-1. Epub 2014 Jan 5. Protoplasma. 2014. PMID: 24390311 Free PMC article. Review.
References
-
- Amat F, Moussavi F, Comolli LR, Elidan G, Downing KH, Horowitz M. Markov random field based automatic image alignment for electron tomography. J. Struct. Biol. 2008;161:260–275. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

