Rapid nucleotide exchange renders Asp-11 mutant actins resistant to depolymerizing activity of cofilin, leading to dominant toxicity in vivo
- PMID: 23212920
- PMCID: PMC3548484
- DOI: 10.1074/jbc.M112.404657
Rapid nucleotide exchange renders Asp-11 mutant actins resistant to depolymerizing activity of cofilin, leading to dominant toxicity in vivo
Abstract
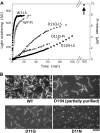
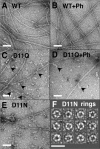
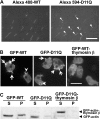
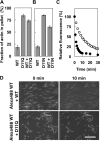
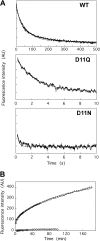
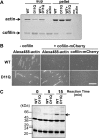
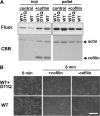
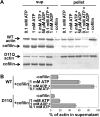
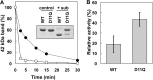
Conserved Asp-11 of actin is a part of the nucleotide binding pocket, and its mutation to Gln is dominant lethal in yeast, whereas the mutation to Asn in human α-actin dominantly causes congenital myopathy. To elucidate the molecular mechanism of those dominant negative effects, we prepared Dictyostelium versions of D11N and D11Q mutant actins and characterized them in vitro. D11N and D11Q actins underwent salt-dependent reversible polymerization, although the resultant polymerization products contained small anomalous structures in addition to filaments of normal appearance. Both monomeric and polymeric D11Q actin released bound nucleotides more rapidly than the wild type, and intriguingly, both monomeric and polymeric D11Q actins hardly bound cofilin. The deficiency in cofilin binding can be explained by rapid exchange of bound nucleotide with ATP in solution, because cofilin does not bind ATP-bound actin. Copolymers of D11Q and wild type actins bound cofilin, but cofilin-induced depolymerization of the copolymers was slower than that of wild type filaments, which may presumably be the primary reason why this mutant actin is dominantly toxic in vivo. Purified D11N actin was unstable, which made its quantitative biochemical characterization difficult. However, monomeric D11N actin released nucleotides even faster than D11Q, and we speculate that D11N actin also exerts its toxic effects in vivo through a defective interaction with cofilin. We have recently found that two other dominant negative actin mutants are also defective in cofilin binding, and we propose that the defective cofilin binder is a major class of dominant negative actin mutants.
Figures
Similar articles
-
Cofilin-induced cooperative conformational changes of actin subunits revealed using cofilin-actin fusion protein.Sci Rep. 2016 Feb 4;6:20406. doi: 10.1038/srep20406. Sci Rep. 2016. PMID: 26842224 Free PMC article.
-
Screening of novel dominant negative mutant actins using glycine targeted scanning identifies G146V actin that cooperatively inhibits cofilin binding.Biochem Biophys Res Commun. 2010 Jun 11;396(4):1006-11. doi: 10.1016/j.bbrc.2010.05.047. Epub 2010 May 13. Biochem Biophys Res Commun. 2010. PMID: 20471369
-
K336I mutant actin alters the structure of neighbouring protomers in filaments and reduces affinity for actin-binding proteins.Sci Rep. 2019 Mar 29;9(1):5353. doi: 10.1038/s41598-019-41795-w. Sci Rep. 2019. PMID: 30926871 Free PMC article.
-
Cofilin - a protein controlling dynamics of actin filaments.Postepy Hig Med Dosw (Online). 2017 May 5;71(0):339-351. doi: 10.5604/01.3001.0010.3818. Postepy Hig Med Dosw (Online). 2017. PMID: 28513458 Review.
-
Biophysics of actin filament severing by cofilin.FEBS Lett. 2013 Apr 17;587(8):1215-9. doi: 10.1016/j.febslet.2013.01.062. Epub 2013 Feb 5. FEBS Lett. 2013. PMID: 23395798 Free PMC article. Review.
Cited by
-
Cofilin-induced changes in F-actin detected via cross-linking with benzophenone-4-maleimide.Biochemistry. 2013 Aug 13;52(32):5503-9. doi: 10.1021/bi400715z. Epub 2013 Jul 31. Biochemistry. 2013. PMID: 23862734 Free PMC article.
-
Allosteric regulation by cooperative conformational changes of actin filaments drives mutually exclusive binding with cofilin and myosin.Sci Rep. 2016 Oct 20;6:35449. doi: 10.1038/srep35449. Sci Rep. 2016. PMID: 27762277 Free PMC article.
-
Prevalence of Cytoplasmic Actin Mutations in Diffuse Large B-Cell Lymphoma and Multiple Myeloma: A Functional Assessment Based on Actin Three-Dimensional Structures.Int J Mol Sci. 2020 Apr 27;21(9):3093. doi: 10.3390/ijms21093093. Int J Mol Sci. 2020. PMID: 32349449 Free PMC article.
-
Cofilin-induced cooperative conformational changes of actin subunits revealed using cofilin-actin fusion protein.Sci Rep. 2016 Feb 4;6:20406. doi: 10.1038/srep20406. Sci Rep. 2016. PMID: 26842224 Free PMC article.
-
ATP-dependent regulation of actin monomer-filament equilibrium by cyclase-associated protein and ADF/cofilin.Biochem J. 2013 Jul 15;453(2):249-59. doi: 10.1042/BJ20130491. Biochem J. 2013. PMID: 23672398 Free PMC article.
References
-
- An H. S., Mogami K. (1996) Isolation of 88F actin mutants of Drosophila melanogaster and possible alterations in the mutant actin structures. J. Mol. Biol. 260, 492–505 - PubMed
-
- Noguchi T. Q., Toya R., Ueno H., Tokuraku K., Uyeda T. Q. (2010) Screening of novel dominant negative mutant actins using glycine targeted scanning identifies G146V actin that cooperatively inhibits cofilin binding. Biochem. Biophys. Res. Commun. 396, 1006–1011 - PubMed
-
- Drummond D. R., Hennessey E. S., Sparrow J. C. (1991) Characterisation of missense mutations in the Act88F gene of. Drosophila melanogaster. Mol. Gen. Genet. 226, 70–80 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

