Coronavirus genotype diversity and prevalence of infection in wild carnivores in the Serengeti National Park, Tanzania
- PMID: 23212740
- PMCID: PMC7086904
- DOI: 10.1007/s00705-012-1562-x
Coronavirus genotype diversity and prevalence of infection in wild carnivores in the Serengeti National Park, Tanzania
Abstract
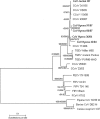
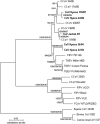
Knowledge of coronaviruses in wild carnivores is limited. This report describes coronavirus genetic diversity, species specificity and infection prevalence in three wild African carnivores. Coronavirus RNA was recovered from fresh feces from spotted hyena and silver-backed jackal, but not bat-eared fox. Analysis of sequences of membrane (M) and spike (S) gene fragments revealed strains in the genus Alphacoronavirus, including three distinct strains in hyenas and one distinct strain in a jackal. Coronavirus RNA prevalence was higher in feces from younger (17 %) than older (3 %) hyenas, highlighting the importance of young animals for coronavirus transmission in wild carnivores.
Figures
Similar articles
-
Divergent Sapovirus Strains and Infection Prevalence in Wild Carnivores in the Serengeti Ecosystem: A Long-Term Study.PLoS One. 2016 Sep 23;11(9):e0163548. doi: 10.1371/journal.pone.0163548. eCollection 2016. PLoS One. 2016. PMID: 27661997 Free PMC article.
-
Coronavirus infection of spotted hyenas in the Serengeti ecosystem.Vet Microbiol. 2004 Aug 19;102(1-2):1-9. doi: 10.1016/j.vetmic.2004.04.012. Vet Microbiol. 2004. PMID: 15288921 Free PMC article.
-
The virus-host interface: Molecular interactions of Alphacoronavirus-1 variants from wild and domestic hosts with mammalian aminopeptidase N.Mol Ecol. 2021 Jun;30(11):2607-2625. doi: 10.1111/mec.15910. Epub 2021 May 3. Mol Ecol. 2021. PMID: 33786949 Free PMC article.
-
Molecular characterization of canine kobuvirus in wild carnivores and the domestic dog in Africa.Virology. 2015 Mar;477:89-97. doi: 10.1016/j.virol.2015.01.010. Epub 2015 Feb 7. Virology. 2015. PMID: 25667111
-
Genomic and serological detection of bat coronavirus from bats in the Philippines.Arch Virol. 2012 Dec;157(12):2349-55. doi: 10.1007/s00705-012-1410-z. Epub 2012 Jul 26. Arch Virol. 2012. PMID: 22833101 Free PMC article.
Cited by
-
Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses.Vet Microbiol. 2020 May;244:108693. doi: 10.1016/j.vetmic.2020.108693. Epub 2020 Apr 14. Vet Microbiol. 2020. PMID: 32402329 Free PMC article. Review.
-
Absence of Hepatitis E Virus (HEV) Circulation in the Most Widespread Wild Croatian Canine Species, the Red Fox (Vulpes vulpes) and Jackal (Canis aureus moreoticus).Microorganisms. 2023 Mar 24;11(4):834. doi: 10.3390/microorganisms11040834. Microorganisms. 2023. PMID: 37110256 Free PMC article.
-
Divergent Sapovirus Strains and Infection Prevalence in Wild Carnivores in the Serengeti Ecosystem: A Long-Term Study.PLoS One. 2016 Sep 23;11(9):e0163548. doi: 10.1371/journal.pone.0163548. eCollection 2016. PLoS One. 2016. PMID: 27661997 Free PMC article.
-
Comparative survey of canine parvovirus, canine distemper virus and canine enteric coronavirus infection in free-ranging wolves of central Italy and south-eastern France.Eur J Wildl Res. 2014;60(4):613-624. doi: 10.1007/s10344-014-0825-0. Epub 2014 Jun 24. Eur J Wildl Res. 2014. PMID: 32214941 Free PMC article.
-
Social status mediates the fitness costs of infection with canine distemper virus in Serengeti spotted hyenas.Funct Ecol. 2018 May;32(5):1237-1250. doi: 10.1111/1365-2435.13059. Epub 2018 Mar 6. Funct Ecol. 2018. PMID: 32313354 Free PMC article.
References
-
- Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Oxford: Oxford University Press; 1991.
-
- Dong BQ, Liu W, Fan XH, Vijaykrishna D, Tang XC, Gao F, Li LF, Li GJ, Zhang JX, Peiris JSM, Smith GJD, Chen H, Guan Y. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in southern China. J Virol. 2007;81:6920–6926. doi: 10.1128/JVI.00299-07. - DOI - PMC - PubMed
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

