Type IV pilin proteins: versatile molecular modules
- PMID: 23204365
- PMCID: PMC3510520
- DOI: 10.1128/MMBR.00035-12
Type IV pilin proteins: versatile molecular modules
Abstract
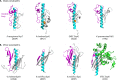
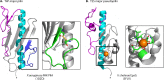
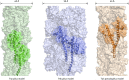
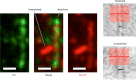
Type IV pili (T4P) are multifunctional protein fibers produced on the surfaces of a wide variety of bacteria and archaea. The major subunit of T4P is the type IV pilin, and structurally related proteins are found as components of the type II secretion (T2S) system, where they are called pseudopilins; of DNA uptake/competence systems in both Gram-negative and Gram-positive species; and of flagella, pili, and sugar-binding systems in the archaea. This broad distribution of a single protein family implies both a common evolutionary origin and a highly adaptable functional plan. The type IV pilin is a remarkably versatile architectural module that has been adopted widely for a variety of functions, including motility, attachment to chemically diverse surfaces, electrical conductance, acquisition of DNA, and secretion of a broad range of structurally distinct protein substrates. In this review, we consider recent advances in this research area, from structural revelations to insights into diversity, posttranslational modifications, regulation, and function.
Figures



Similar articles
-
Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin.J Biol Chem. 2015 Jan 2;290(1):601-11. doi: 10.1074/jbc.M114.616904. Epub 2014 Nov 11. J Biol Chem. 2015. PMID: 25389296 Free PMC article.
-
The Biosynthesis and Structures of Bacterial Pili.Subcell Biochem. 2019;92:369-413. doi: 10.1007/978-3-030-18768-2_12. Subcell Biochem. 2019. PMID: 31214993 Review.
-
Bridging Bacteria and the Gut: Functional Aspects of Type IV Pili.Trends Microbiol. 2020 May;28(5):340-348. doi: 10.1016/j.tim.2020.02.003. Epub 2020 Mar 5. Trends Microbiol. 2020. PMID: 32298612 Review.
-
Structure of the competence pilus major pilin ComGC in Streptococcus pneumoniae.J Biol Chem. 2017 Aug 25;292(34):14134-14146. doi: 10.1074/jbc.M117.787671. Epub 2017 Jun 28. J Biol Chem. 2017. PMID: 28659339 Free PMC article.
-
Type IV pili: dynamics, biophysics and functional consequences.Nat Rev Microbiol. 2019 Jul;17(7):429-440. doi: 10.1038/s41579-019-0195-4. Nat Rev Microbiol. 2019. PMID: 30988511 Review.
Cited by
-
Type IV Pili Are a Critical Virulence Factor in Clinical Isolates of Paenibacillus thiaminolyticus.mBio. 2022 Dec 20;13(6):e0268822. doi: 10.1128/mbio.02688-22. Epub 2022 Nov 14. mBio. 2022. PMID: 36374038 Free PMC article.
-
High-Throughput Screen for Inhibitors of the Type IV Pilus Assembly ATPase PilB.mSphere. 2021 Mar 3;6(2):e00129-21. doi: 10.1128/mSphere.00129-21. mSphere. 2021. PMID: 33658276 Free PMC article.
-
Structural Diversity in the Type IV Pili of Multidrug-resistant Acinetobacter.J Biol Chem. 2016 Oct 28;291(44):22924-22935. doi: 10.1074/jbc.M116.751099. Epub 2016 Sep 15. J Biol Chem. 2016. PMID: 27634041 Free PMC article.
-
Evaluation of the Immunoprotective Capacity of Five Vaccine Candidate Proteins against Avian Necrotic Enteritis and Impact on the Caecal Microbiota of Vaccinated Birds.Animals (Basel). 2023 Oct 26;13(21):3323. doi: 10.3390/ani13213323. Animals (Basel). 2023. PMID: 37958078 Free PMC article.
-
FlaF Is a β-Sandwich Protein that Anchors the Archaellum in the Archaeal Cell Envelope by Binding the S-Layer Protein.Structure. 2015 May 5;23(5):863-872. doi: 10.1016/j.str.2015.03.001. Epub 2015 Apr 9. Structure. 2015. PMID: 25865246 Free PMC article.
References
-
- Aas FE, et al. 2006. Neisseria gonorrhoeae type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J. Biol. Chem. 281: 27712–27723 - PubMed
-
- Aas FE, Lovold C, Koomey M. 2002. An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to type IV pilus expression. Mol. Microbiol. 46: 1441–1450 - PubMed
-
- Aas FE, et al. 2007. Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect type IV pilus assembly, dynamics and associated functions. Mol. Microbiol. 63: 69–85 - PubMed
-
- Aas FE, et al. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46: 749–760 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

