The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation
- PMID: 23202476
- PMCID: PMC3619131
- DOI: 10.1016/j.bbagrm.2012.11.005
The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation
Abstract
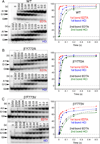
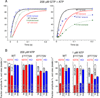
The bridge α-helix in the β' subunit of RNA polymerase (RNAP) borders the active site and may have roles in catalysis and translocation. In Escherichia coli RNAP, a bulky hydrophobic segment near the N-terminal end of the bridge helix is identified (β' 772-YFI-774; the YFI motif). YFI is located at a distance from the active center and adjacent to a glycine hinge (β' 778-GARKG-782) involved in dynamic bending of the bridge helix. Remarkably, amino acid substitutions in YFI significantly alter intrinsic termination, pausing, fidelity and translocation of RNAP. F773V RNAP largely ignores the λ tR2 terminator at 200μM NTPs and is strongly reduced in λ tR2 recognition at 1μM NTPs. F773V alters RNAP pausing and backtracking and favors misincorporation. By contrast, the adjacent Y772A substitution increases fidelity and exhibits other transcriptional defects generally opposite to those of F773V. All atom molecular dynamics simulation revealed two separate functional connections emanating from YFI explaining the distinct effects of substitutions: Y772 communicates with the active site through the link domain in the β subunit, whereas F773 communicates through the fork domain in the β subunit. I774 interacts with the F-loop, which also contacts the glycine hinge of the bridge helix. These results identified negative and positive circuits coupled at YFI and employed for regulation of catalysis, elongation, termination and translocation.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures






Similar articles
-
Hinge action versus grip in translocation by RNA polymerase.Transcription. 2018;9(1):1-16. doi: 10.1080/21541264.2017.1330179. Epub 2017 Aug 30. Transcription. 2018. PMID: 28853995 Free PMC article.
-
Conformational coupling, bridge helix dynamics and active site dehydration in catalysis by RNA polymerase.Biochim Biophys Acta. 2010 Aug;1799(8):575-87. doi: 10.1016/j.bbagrm.2010.05.002. Epub 2010 May 15. Biochim Biophys Acta. 2010. PMID: 20478425 Free PMC article.
-
Molecular dynamics and mutational analysis of the catalytic and translocation cycle of RNA polymerase.BMC Biophys. 2012 Jun 7;5:11. doi: 10.1186/2046-1682-5-11. BMC Biophys. 2012. PMID: 22676913 Free PMC article.
-
The Bridge Helix of RNA polymerase acts as a central nanomechanical switchboard for coordinating catalysis and substrate movement.Archaea. 2011;2011:608385. doi: 10.1155/2011/608385. Epub 2012 Jan 22. Archaea. 2011. PMID: 22312317 Free PMC article. Review.
-
Structural perspective on mutations affecting the function of multisubunit RNA polymerases.Microbiol Mol Biol Rev. 2006 Mar;70(1):12-36. doi: 10.1128/MMBR.70.1.12-36.2006. Microbiol Mol Biol Rev. 2006. PMID: 16524917 Free PMC article. Review.
Cited by
-
Active site closure stabilizes the backtracked state of RNA polymerase.Nucleic Acids Res. 2018 Nov 16;46(20):10870-10887. doi: 10.1093/nar/gky883. Nucleic Acids Res. 2018. PMID: 30256972 Free PMC article.
-
Computational simulation strategies for analysis of multisubunit RNA polymerases.Chem Rev. 2013 Nov 13;113(11):8546-66. doi: 10.1021/cr400046x. Epub 2013 Aug 29. Chem Rev. 2013. PMID: 23987500 Free PMC article. Review. No abstract available.
-
Transcription-translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits.Nucleic Acids Res. 2017 Nov 2;45(19):11043-11055. doi: 10.1093/nar/gkx719. Nucleic Acids Res. 2017. PMID: 28977553 Free PMC article.
-
Millisecond dynamics of RNA polymerase II translocation at atomic resolution.Proc Natl Acad Sci U S A. 2014 May 27;111(21):7665-70. doi: 10.1073/pnas.1315751111. Epub 2014 Apr 21. Proc Natl Acad Sci U S A. 2014. PMID: 24753580 Free PMC article.
-
Translocation and fidelity of Escherichia coli RNA polymerase.Transcription. 2013 May-Jun;4(3):136-43. doi: 10.4161/trns.25527. Epub 2013 Jul 11. Transcription. 2013. PMID: 23863783 Free PMC article.
References
-
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

