Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease
- PMID: 23201127
- PMCID: PMC3586414
- DOI: 10.1016/j.molcel.2012.10.023
Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease
Abstract
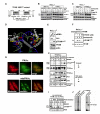
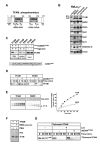
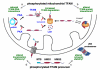
Human mitochondrial transcription factor A (TFAM) is a high-mobility group (HMG) protein at the nexus of mitochondrial DNA (mtDNA) replication, transcription, and inheritance. Little is known about the mechanisms underlying its posttranslational regulation. Here, we demonstrate that TFAM is phosphorylated within its HMG box 1 (HMG1) by cAMP-dependent protein kinase in mitochondria. HMG1 phosphorylation impairs the ability of TFAM to bind DNA and to activate transcription. We show that only DNA-free TFAM is degraded by the Lon protease, which is inhibited by the anticancer drug bortezomib. In cells with normal mtDNA levels, HMG1-phosphorylated TFAM is degraded by Lon. However, in cells with severe mtDNA deficits, nonphosphorylated TFAM is also degraded, as it is DNA free. Depleting Lon in these cells increases levels of TFAM and upregulates mtDNA content, albeit transiently. Phosphorylation and proteolysis thus provide mechanisms for rapid fine-tuning of TFAM function and abundance in mitochondria, which are crucial for maintaining and expressing mtDNA.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM).Proc Natl Acad Sci U S A. 2010 Oct 26;107(43):18410-5. doi: 10.1073/pnas.1008924107. Epub 2010 Oct 7. Proc Natl Acad Sci U S A. 2010. PMID: 20930118 Free PMC article.
-
Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA.Mol Cell Biol. 2004 Nov;24(22):9823-34. doi: 10.1128/MCB.24.22.9823-9834.2004. Mol Cell Biol. 2004. PMID: 15509786 Free PMC article.
-
Tetramethylpyrazine blocks TFAM degradation and up-regulates mitochondrial DNA copy number by interacting with TFAM.Biosci Rep. 2017 May 17;37(3):BSR20170319. doi: 10.1042/BSR20170319. Print 2017 Jun 30. Biosci Rep. 2017. PMID: 28465355 Free PMC article.
-
Matrix proteases in mitochondrial DNA function.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):1080-7. doi: 10.1016/j.bbagrm.2011.11.008. Epub 2011 Dec 8. Biochim Biophys Acta. 2012. PMID: 22172992 Free PMC article. Review.
-
Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):921-9. doi: 10.1016/j.bbagrm.2012.03.002. Epub 2012 Mar 21. Biochim Biophys Acta. 2012. PMID: 22465614 Review.
Cited by
-
The cAMP phosphodiesterase Prune localizes to the mitochondrial matrix and promotes mtDNA replication by stabilizing TFAM.EMBO Rep. 2015 Apr;16(4):520-7. doi: 10.15252/embr.201439636. Epub 2015 Feb 3. EMBO Rep. 2015. PMID: 25648146 Free PMC article.
-
Powering down the mitochondrial LonP1 protease: a novel strategy for anticancer therapeutics.Expert Opin Ther Targets. 2024 Jan-Feb;28(1-2):9-15. doi: 10.1080/14728222.2023.2298358. Epub 2023 Dec 29. Expert Opin Ther Targets. 2024. PMID: 38156441 Free PMC article. Review.
-
Molecular Determinants of Mitochondrial Shape and Function and Their Role in Glaucoma.Antioxid Redox Signal. 2023 May;38(13-15):896-919. doi: 10.1089/ars.2022.0124. Epub 2023 Jan 5. Antioxid Redox Signal. 2023. PMID: 36301938 Free PMC article. Review.
-
LONP1 Is Required for Maturation of a Subset of Mitochondrial Proteins, and Its Loss Elicits an Integrated Stress Response.Mol Cell Biol. 2018 Sep 28;38(20):e00412-17. doi: 10.1128/MCB.00412-17. Print 2018 Oct 15. Mol Cell Biol. 2018. PMID: 30061372 Free PMC article.
-
TRPC6-Mediated ERK1/2 Activation Increases Dentate Granule Cell Resistance to Status Epilepticus Via Regulating Lon Protease-1 Expression and Mitochondrial Dynamics.Cells. 2019 Nov 1;8(11):1376. doi: 10.3390/cells8111376. Cells. 2019. PMID: 31683954 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

