Asymmetric regulation of bipolar single-stranded DNA translocation by the two motors within Escherichia coli RecBCD helicase
- PMID: 23192341
- PMCID: PMC3542991
- DOI: 10.1074/jbc.M112.423384
Asymmetric regulation of bipolar single-stranded DNA translocation by the two motors within Escherichia coli RecBCD helicase
Abstract
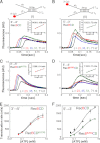
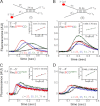
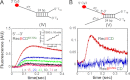
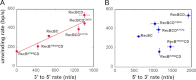
Repair of double-stranded DNA breaks in Escherichia coli is initiated by the RecBCD helicase that possesses two superfamily-1 motors, RecB (3' to 5' translocase) and RecD (5' to 3' translocase), that operate on the complementary DNA strands to unwind duplex DNA. However, it is not known whether the RecB and RecD motors act independently or are functionally coupled. Here we show by directly monitoring ATP-driven single-stranded DNA translocation of RecBCD that the 5' to 3' rate is always faster than the 3' to 5' rate on DNA without a crossover hotspot instigator site and that the translocation rates are coupled asymmetrically. That is, RecB regulates both 3' to 5' and 5' to 3' translocation, whereas RecD only regulates 5' to 3' translocation. We show that the recently identified RecBC secondary translocase activity functions within RecBCD and that this contributes to the coupling. This coupling has implications for how RecBCD activity is regulated after it recognizes a crossover hotspot instigator sequence during DNA unwinding.
Figures

Similar articles
-
Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme.J Biol Chem. 2005 Nov 4;280(44):37069-77. doi: 10.1074/jbc.M505520200. Epub 2005 Jul 22. J Biol Chem. 2005. PMID: 16041061
-
Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor.Nat Struct Mol Biol. 2010 Oct;17(10):1210-7. doi: 10.1038/nsmb.1901. Epub 2010 Sep 19. Nat Struct Mol Biol. 2010. PMID: 20852646 Free PMC article.
-
E. coli RecBCD Nuclease Domain Regulates Helicase Activity but not Single Stranded DNA Translocase Activity.bioRxiv [Preprint]. 2023 Oct 17:2023.10.13.561901. doi: 10.1101/2023.10.13.561901. bioRxiv. 2023. Update in: J Mol Biol. 2024 Jan 15;436(2):168381. doi: 10.1016/j.jmb.2023.168381. PMID: 37905078 Free PMC article. Updated. Preprint.
-
RecBCD enzyme and the repair of double-stranded DNA breaks.Microbiol Mol Biol Rev. 2008 Dec;72(4):642-71, Table of Contents. doi: 10.1128/MMBR.00020-08. Microbiol Mol Biol Rev. 2008. PMID: 19052323 Free PMC article. Review.
-
Chi and the RecBC D enzyme of Escherichia coli.Annu Rev Genet. 1994;28:49-70. doi: 10.1146/annurev.ge.28.120194.000405. Annu Rev Genet. 1994. PMID: 7893137 Review.
Cited by
-
E. coli RecB Nuclease Domain Regulates RecBCD Helicase Activity but not Single Stranded DNA Translocase Activity.J Mol Biol. 2024 Jan 15;436(2):168381. doi: 10.1016/j.jmb.2023.168381. Epub 2023 Dec 9. J Mol Biol. 2024. PMID: 38081382 Free PMC article.
-
A new twist on PIFE: photoisomerisation-related fluorescence enhancement.ArXiv [Preprint]. 2023 Jul 10:arXiv:2302.12455v2. ArXiv. 2023. Update in: Methods Appl Fluoresc. 2023 Oct 12;12(1). doi: 10.1088/2050-6120/acfb58. PMID: 36866225 Free PMC article. Updated. Preprint.
-
Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting.J Mol Biol. 2021 Sep 3;433(18):167147. doi: 10.1016/j.jmb.2021.167147. Epub 2021 Jul 9. J Mol Biol. 2021. PMID: 34246654 Free PMC article.
-
Synergy between RecBCD subunits is essential for efficient DNA unwinding.Elife. 2019 Jan 2;8:e40836. doi: 10.7554/eLife.40836. Elife. 2019. PMID: 30601118 Free PMC article.
-
Processive DNA Unwinding by RecBCD Helicase in the Absence of Canonical Motor Translocation.J Mol Biol. 2016 Jul 31;428(15):2997-3012. doi: 10.1016/j.jmb.2016.07.002. Epub 2016 Jul 14. J Mol Biol. 2016. PMID: 27422010 Free PMC article.
References
-
- Anderson D. G., Kowalczykowski S. C. (1997) The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell 90, 77–86 - PubMed
-
- Smith G. R. (1989) Homologous recombination in prokaryotes: enzymes and controlling sites. Genome 31, 520–527 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

