Expression analysis of inflammasomes in experimental models of inflammatory and fibrotic liver disease
- PMID: 23192004
- PMCID: PMC3599703
- DOI: 10.1186/1476-9255-9-49
Expression analysis of inflammasomes in experimental models of inflammatory and fibrotic liver disease
Abstract
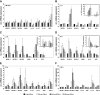
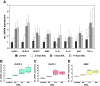
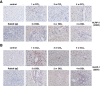
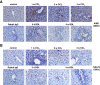
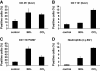
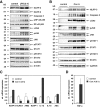
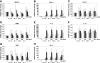
During inflammation, the inflammasomes representing a group of multi-protein complexes trigger the biological maturation of pro-inflammatory cytokines such as interleukin-1β and interleukin-18 by proteolytic activation of caspase-1 from its inactive proforms. The individual genes encoding components of the inflammasome machinery are regulated at transcriptional and post-transcriptional levels. Once activated, they drive a wide variety of cellular responses that are necessary to mediate host defense against microbial pathogens and to guarantee tissue homeostasis. In the present work, we have studied the expression of the different inflammasomes in various primary hepatic cell subpopulations, in models of acute inflammation and during experimental liver fibrogenesis. We demonstrate that NLRP-1, NLRP-3 and AIM2 are prominently expressed in Kupffer cells and liver sinusoidal endothelial cells, moderately expressed in periportal myofibroblasts and hepatic stellate cells, and virtually absent in primary cultured hepatocytes. We found that the challenge with the lipopolysaccharides results in a time- and concentration-dependent expression of the NOD-like receptor family members NLRP-1, NLRP-3 and NLRC4/NALP4 in cultured hepatic stellate cells and a strong transcriptional activation of NLRP-3 in hepatocytes. Moreover, we detect a diverse regulatory network of the different inflammasomes in the chosen experimental models of acute and chronic liver insult suggesting that the various inflammasomes might contribute simultaneously to the outcome of inflammatory and fibrotic liver insult, irrespectively of the underlying inflammatory stimulus.
Figures
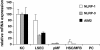

Similar articles
-
Cooperation of liver cells in health and disease.Adv Anat Embryol Cell Biol. 2001;161:III-XIII, 1-151. doi: 10.1007/978-3-642-56553-3. Adv Anat Embryol Cell Biol. 2001. PMID: 11729749 Review.
-
NLRP3 inflammasome expression is driven by NF-κB in cultured hepatocytes.Biochem Biophys Res Commun. 2015 Mar 13;458(3):700-706. doi: 10.1016/j.bbrc.2015.02.029. Epub 2015 Feb 14. Biochem Biophys Res Commun. 2015. PMID: 25686493
-
Inflammasomes in intestinal inflammation and cancer.Gastroenterology. 2011 Dec;141(6):1986-99. doi: 10.1053/j.gastro.2011.10.002. Epub 2011 Oct 15. Gastroenterology. 2011. PMID: 22005480 Free PMC article. Review.
-
Cytokine Secretion and Pyroptosis of Thyroid Follicular Cells Mediated by Enhanced NLRP3, NLRP1, NLRC4, and AIM2 Inflammasomes Are Associated With Autoimmune Thyroiditis.Front Immunol. 2018 Jun 4;9:1197. doi: 10.3389/fimmu.2018.01197. eCollection 2018. Front Immunol. 2018. PMID: 29915579 Free PMC article.
-
Regulation and Function of the Nucleotide Binding Domain Leucine-Rich Repeat-Containing Receptor, Pyrin Domain-Containing-3 Inflammasome in Lung Disease.Am J Respir Cell Mol Biol. 2016 Feb;54(2):151-60. doi: 10.1165/rcmb.2015-0231TR. Am J Respir Cell Mol Biol. 2016. PMID: 26418144 Free PMC article. Review.
Cited by
-
Cellular and molecular functions of hepatic stellate cells in inflammatory responses and liver immunology.Hepatobiliary Surg Nutr. 2014 Dec;3(6):344-63. doi: 10.3978/j.issn.2304-3881.2014.11.03. Hepatobiliary Surg Nutr. 2014. PMID: 25568859 Free PMC article. Review.
-
Dual CTLA-4 and PD-L1 Blockade Inhibits Tumor Growth and Liver Metastasis in a Highly Aggressive Orthotopic Mouse Model of Colon Cancer.Neoplasia. 2019 Sep;21(9):932-944. doi: 10.1016/j.neo.2019.07.006. Epub 2019 Aug 11. Neoplasia. 2019. PMID: 31412307 Free PMC article.
-
Molecular Targets in Hepatocarcinogenesis and Implications for Therapy.J Clin Med. 2018 Aug 13;7(8):213. doi: 10.3390/jcm7080213. J Clin Med. 2018. PMID: 30104473 Free PMC article. Review.
-
Alcohol dysregulates miR-148a in hepatocytes through FoxO1, facilitating pyroptosis via TXNIP overexpression.Gut. 2019 Apr;68(4):708-720. doi: 10.1136/gutjnl-2017-315123. Epub 2018 Feb 23. Gut. 2019. PMID: 29475852 Free PMC article.
-
Liver Sinusoidal Endothelial Cell: An Update.Semin Liver Dis. 2017 Nov;37(4):377-387. doi: 10.1055/s-0037-1617455. Epub 2017 Dec 22. Semin Liver Dis. 2017. PMID: 29272898 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

