IL-13-induced airway mucus production is attenuated by MAPK13 inhibition
- PMID: 23187130
- PMCID: PMC3533556
- DOI: 10.1172/JCI64896
IL-13-induced airway mucus production is attenuated by MAPK13 inhibition
Abstract
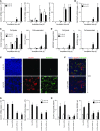
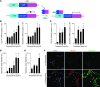
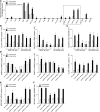
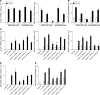
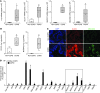
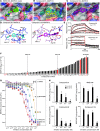
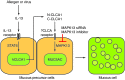
Increased mucus production is a common cause of morbidity and mortality in inflammatory airway diseases, including asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis. However, the precise molecular mechanisms for pathogenic mucus production are largely undetermined. Accordingly, there are no specific and effective anti-mucus therapeutics. Here, we define a signaling pathway from chloride channel calcium-activated 1 (CLCA1) to MAPK13 that is responsible for IL-13-driven mucus production in human airway epithelial cells. The same pathway was also highly activated in the lungs of humans with excess mucus production due to COPD. We further validated the pathway by using structure-based drug design to develop a series of novel MAPK13 inhibitors with nanomolar potency that effectively reduced mucus production in human airway epithelial cells. These results uncover and validate a new pathway for regulating mucus production as well as a corresponding therapeutic approach to mucus overproduction in inflammatory airway diseases.
Figures
Similar articles
-
Increased human Ca²⁺-activated Cl⁻ channel 1 expression and mucus overproduction in airway epithelia of smokers and chronic obstructive pulmonary disease patients.Respir Res. 2012 Jun 25;13(1):55. doi: 10.1186/1465-9921-13-55. Respir Res. 2012. PMID: 22731784 Free PMC article.
-
New pharmacotherapy for airway mucus hypersecretion in asthma and COPD: targeting intracellular signaling pathways.J Aerosol Med Pulm Drug Deliv. 2010 Aug;23(4):219-31. doi: 10.1089/jamp.2009.0802. J Aerosol Med Pulm Drug Deliv. 2010. PMID: 20695774 Review.
-
TMEM16A Mediates Mucus Production in Human Airway Epithelial Cells.Am J Respir Cell Mol Biol. 2021 Jan;64(1):50-58. doi: 10.1165/rcmb.2019-0442OC. Am J Respir Cell Mol Biol. 2021. PMID: 33026825
-
Airway mucus: the good, the bad, the sticky.Pharmacol Ther. 2009 Mar;121(3):332-48. doi: 10.1016/j.pharmthera.2008.11.001. Epub 2008 Nov 18. Pharmacol Ther. 2009. PMID: 19059283 Free PMC article. Review.
-
Treatment of airway mucus hypersecretion.Ann Med. 2006;38(2):116-25. doi: 10.1080/07853890600585795. Ann Med. 2006. PMID: 16581697 Review.
Cited by
-
Group 2 Innate Lymphoid Cells Must Partner with the Myeloid-Macrophage Lineage for Long-Term Postviral Lung Disease.J Immunol. 2020 Aug 15;205(4):1084-1101. doi: 10.4049/jimmunol.2000181. Epub 2020 Jul 8. J Immunol. 2020. PMID: 32641386 Free PMC article.
-
Prioritizing Molecular Biomarkers in Asthma and Respiratory Allergy Using Systems Biology.Front Immunol. 2021 Apr 15;12:640791. doi: 10.3389/fimmu.2021.640791. eCollection 2021. Front Immunol. 2021. PMID: 33936056 Free PMC article.
-
Targeting cell signaling in allergic asthma.Signal Transduct Target Ther. 2019 Oct 18;4:45. doi: 10.1038/s41392-019-0079-0. eCollection 2019. Signal Transduct Target Ther. 2019. PMID: 31637021 Free PMC article. Review.
-
Solithromycin inhibits IL-13-induced goblet cell hyperplasia and MUC5AC, CLCA1, and ANO1 in human bronchial epithelial cells.PeerJ. 2023 Jan 17;11:e14695. doi: 10.7717/peerj.14695. eCollection 2023. PeerJ. 2023. PMID: 36684665 Free PMC article.
-
Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase.Nat Commun. 2019 Jan 10;10(1):131. doi: 10.1038/s41467-018-08027-7. Nat Commun. 2019. PMID: 30631068 Free PMC article.
References
-
- Kuyper LM, et al. Characterization of airway plugging in fatal asthma. Am J Med. 2003;115(1):6–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL029594/HL/NHLBI NIH HHS/United States
- K08 HL083095/HL/NHLBI NIH HHS/United States
- P30AR048335/AR/NIAMS NIH HHS/United States
- P50-HL107183/HL/NHLBI NIH HHS/United States
- U19 AI070489/AI/NIAID NIH HHS/United States
- P50 HL107183/HL/NHLBI NIH HHS/United States
- U19-AI070489/AI/NIAID NIH HHS/United States
- P30 DC004665/DC/NIDCD NIH HHS/United States
- R01 HL073159/HL/NHLBI NIH HHS/United States
- P30DC04665/DC/NIDCD NIH HHS/United States
- R01-HL073159/HL/NHLBI NIH HHS/United States
- K08-HL083095/HL/NHLBI NIH HHS/United States
- P30 AR048335/AR/NIAMS NIH HHS/United States
- P01-HL29594/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

