Differential roles of fibroblast growth factor receptors (FGFR) 1, 2 and 3 in the regulation of S115 breast cancer cell growth
- PMID: 23185502
- PMCID: PMC3503871
- DOI: 10.1371/journal.pone.0049970
Differential roles of fibroblast growth factor receptors (FGFR) 1, 2 and 3 in the regulation of S115 breast cancer cell growth
Abstract
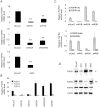
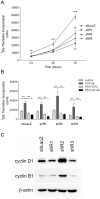
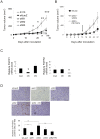
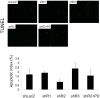
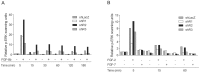
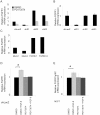
Fibroblast growth factors (FGFs) regulate the growth and progression of breast cancer. FGF signaling is transduced through FGF receptors 1-4, which have oncogenic or anti-oncogenic roles depending on the ligand and the cellular context. Our aim was to clarify the roles of FGFR1-3 in breast cancer cell growth in vitro and in vivo. Pools of S115 mouse breast cancer cells expressing shRNA against FGFR1, 2 and 3 were created by lentiviral gene transfer, resulting in cells with downregulated expression of FGFR1, FGFR2 or FGFR3 (shR1, shR2 and shR3 cells, respectively) and shLacZ controls. FGFR1-silenced shR1 cells formed small, poorly vascularized tumors in nude mice. Silencing of FGFR2 in shR2 cells was associated with strong upregulation of FGFR1 expression and the formation of large, highly vascularized tumors compared to the control tumors. Silencing FGFR3 did not affect cell survival or tumor growth. Overexpressing FGFR2 in control cells did not affect FGFR1 expression, suggesting that high FGFR1 expression in shR2 cells and tumors was associated with FGFR2 silencing by indirect mechanisms. The expression of FGFR1 was, however, increased by the addition of FGF-8 to starved shLacZ or MCF-7 cells and decreased by the FGFR inhibitor PD173074 in shR2 cells with an elevated FGFR1 level. In conclusion, our results demonstrate that FGFR1 is crucial for S115 breast cancer cell proliferation and tumor growth and angiogenesis, whereas FGFR2 and FGFR3 are less critical for the growth of these cells. The results also suggest that the expression of FGFR1 itself is regulated by FGF-8 and FGF signaling, which may be of importance in breast tumors expressing FGFs at a high level.
Conflict of interest statement
Figures


Similar articles
-
Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer.Clin Cancer Res. 2013 Jul 1;19(13):3693-702. doi: 10.1158/1078-0432.CCR-13-0190. Epub 2013 May 8. Clin Cancer Res. 2013. PMID: 23658459 Clinical Trial.
-
Fibroblast growth factor receptor 1 is principally responsible for fibroblast growth factor 2-induced catabolic activities in human articular chondrocytes.Arthritis Res Ther. 2011 Aug 11;13(4):R130. doi: 10.1186/ar3441. Arthritis Res Ther. 2011. Retraction in: Arthritis Res Ther. 2024 Aug 7;26(1):149. doi: 10.1186/s13075-024-03381-y. PMID: 21835001 Free PMC article. Retracted.
-
Selective over-expression of fibroblast growth factor receptors 1 and 4 in clinical prostate cancer.J Pathol. 2007 Sep;213(1):82-90. doi: 10.1002/path.2205. J Pathol. 2007. PMID: 17607666
-
Exploring mechanisms of FGF signalling through the lens of structural biology.Nat Rev Mol Cell Biol. 2013 Mar;14(3):166-80. doi: 10.1038/nrm3528. Epub 2013 Feb 13. Nat Rev Mol Cell Biol. 2013. PMID: 23403721 Free PMC article. Review.
-
FGFR inhibitors: Effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review).Int J Mol Med. 2016 Jul;38(1):3-15. doi: 10.3892/ijmm.2016.2620. Epub 2016 May 31. Int J Mol Med. 2016. PMID: 27245147 Free PMC article. Review.
Cited by
-
Tumor microenvironment regulates metastasis and metastasis genes of mouse MMTV-PymT mammary cancer cells in vivo.Vet Pathol. 2014 Jul;51(4):868-81. doi: 10.1177/0300985813505116. Epub 2013 Oct 3. Vet Pathol. 2014. PMID: 24091811 Free PMC article.
-
The FGFR/MEK/ERK/brachyury pathway is critical for chordoma cell growth and survival.Carcinogenesis. 2014 Jul;35(7):1491-9. doi: 10.1093/carcin/bgu014. Epub 2014 Jan 20. Carcinogenesis. 2014. PMID: 24445144 Free PMC article.
-
2b or Not 2b: How Opposing FGF Receptor Splice Variants Are Blocking Progress in Precision Oncology.J Oncol. 2021 Apr 30;2021:9955456. doi: 10.1155/2021/9955456. eCollection 2021. J Oncol. 2021. PMID: 34007277 Free PMC article. Review.
-
Loss of Dlg-1 in the mouse lens impairs fibroblast growth factor receptor signaling.PLoS One. 2014 May 13;9(5):e97470. doi: 10.1371/journal.pone.0097470. eCollection 2014. PLoS One. 2014. PMID: 24824078 Free PMC article.
-
TNFAIP3 is required for FGFR1 activation-promoted proliferation and tumorigenesis of premalignant DCIS.COM human mammary epithelial cells.Breast Cancer Res. 2018 Aug 15;20(1):97. doi: 10.1186/s13058-018-1024-9. Breast Cancer Res. 2018. PMID: 30111373 Free PMC article.
References
-
- Goldfarb M (1996) Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev 7(4): 311–325. - PubMed
-
- Eswarakumar VP, Lax I, Schlessinger J (2005) Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 16(2): 139–149. - PubMed
-
- Dailey L, Ambrosetti D, Mansukhani A, Basilico C (2005) Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev 16(2): 233–247. - PubMed
-
- Adnane J, Gaudray P, Dionne CA, Crumley G, Jaye M, et al. (1991) BEK and FLG, two receptors to members of the FGF family, are amplified in subsets of human breast cancers. Oncogene 6(4): 659–663. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

