A novel function for SNAP29 (synaptosomal-associated protein of 29 kDa) in mast cell phagocytosis
- PMID: 23185475
- PMCID: PMC3503860
- DOI: 10.1371/journal.pone.0049886
A novel function for SNAP29 (synaptosomal-associated protein of 29 kDa) in mast cell phagocytosis
Abstract
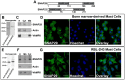
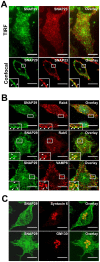
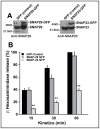
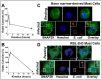
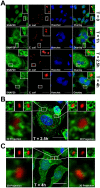
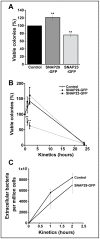
Mast cells play a critical role in the innate immune response to bacterial infection. They internalize and kill a variety of bacteria and process antigen for presentation to T cells via MHC molecules. Although mast cell phagocytosis appears to play a significant role during bacterial infection, little is known about the proteins involved in its regulation. In this study, we demonstrate that the SNARE protein SNAP29 is involved in mast cell phagocytosis. SNAP29 is localized in the endocytic pathway and is transiently recruited to Escherichia coli (E. coli)-containing phagosomes. Interestingly, overexpression of SNAP29 significantly increases the internalization and killing of E. coli, while it does not affect mast cell exocytosis of inflammatory mediators. To our knowledge, these data are the first to demonstrate a novel function of SNAP29 in mast cell phagocytosis and have implications in protection against bacterial infection.
Conflict of interest statement
Figures
Similar articles
-
Allergen-sensitization increases mast-cell expression of the exocytotic proteins SNAP-23 and syntaxin 4, which are involved in histamine secretion.J Investig Allergol Clin Immunol. 2008;18(5):366-71. J Investig Allergol Clin Immunol. 2008. PMID: 18973100
-
Multiple functions of the SNARE protein Snap29 in autophagy, endocytic, and exocytic trafficking during epithelial formation in Drosophila.Autophagy. 2014;10(12):2251-68. doi: 10.4161/15548627.2014.981913. Autophagy. 2014. PMID: 25551675 Free PMC article.
-
SNAP-23 regulates phagosome formation and maturation in macrophages.Mol Biol Cell. 2012 Dec;23(24):4849-63. doi: 10.1091/mbc.E12-01-0069. Epub 2012 Oct 19. Mol Biol Cell. 2012. PMID: 23087210 Free PMC article.
-
Snapshots from within the cell: Novel trafficking and non trafficking functions of Snap29 during tissue morphogenesis.Semin Cell Dev Biol. 2023 Jan 15;133:42-52. doi: 10.1016/j.semcdb.2022.02.024. Epub 2022 Mar 4. Semin Cell Dev Biol. 2023. PMID: 35256275 Review.
-
Mast cell modulation of immune responses to bacteria.Immunol Rev. 2001 Feb;179:16-24. doi: 10.1034/j.1600-065x.2001.790102.x. Immunol Rev. 2001. PMID: 11292019 Review.
Cited by
-
SNARE-Mediated Exocytosis in Neuronal Development.Front Mol Neurosci. 2020 Aug 7;13:133. doi: 10.3389/fnmol.2020.00133. eCollection 2020. Front Mol Neurosci. 2020. PMID: 32848598 Free PMC article. Review.
-
In the line-up: deleted genes associated with DiGeorge/22q11.2 deletion syndrome: are they all suspects?J Neurodev Disord. 2019 Jun 7;11(1):7. doi: 10.1186/s11689-019-9267-z. J Neurodev Disord. 2019. PMID: 31174463 Free PMC article. Review.
-
Role of Mast Cells in clearance of Leishmania through extracellular trap formation.Sci Rep. 2017 Oct 16;7(1):13240. doi: 10.1038/s41598-017-12753-1. Sci Rep. 2017. PMID: 29038500 Free PMC article.
-
An essential step of kinetochore formation controlled by the SNARE protein Snap29.EMBO J. 2016 Oct 17;35(20):2223-2237. doi: 10.15252/embj.201693991. Epub 2016 Sep 19. EMBO J. 2016. PMID: 27647876 Free PMC article.
-
Drosophila SNAP-29 is an essential SNARE that binds multiple proteins involved in membrane traffic.PLoS One. 2014 Mar 13;9(3):e91471. doi: 10.1371/journal.pone.0091471. eCollection 2014. PLoS One. 2014. PMID: 24626111 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

