Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection
- PMID: 23184194
- PMCID: PMC4901162
- DOI: 10.1007/s00018-012-1213-0
Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection
Abstract
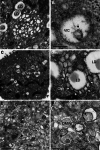
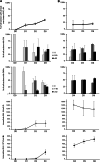
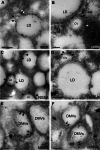
Like most positive-strand RNA viruses, hepatitis C virus (HCV) forms a membrane-associated replication complex consisting of replicating RNA, viral and host proteins anchored to altered cell membranes. We used a combination of qualitative and quantitative electron microscopy (EM), immuno-EM, and the 3D reconstruction of serial EM sections to analyze the host cell membrane alterations induced by HCV. Three different types of membrane alteration were observed: vesicles in clusters (ViCs), contiguous vesicles (CVs), and double-membrane vesicles (DMVs). The main ultrastructural change observed early in infection was the formation of a network of CVs surrounding the lipid droplets. Later stages in the infectious cycle were characterized by a large increase in the number of DMVs, which may be derived from the CVs. These DMVs are thought to constitute the membranous structures harboring the viral replication complexes in which viral replication is firmly and permanently established and to protect the virus against double-stranded RNA-triggered host antiviral responses.
Figures
Similar articles
-
Double-Membrane Vesicles as Platforms for Viral Replication.Trends Microbiol. 2020 Dec;28(12):1022-1033. doi: 10.1016/j.tim.2020.05.009. Epub 2020 Jun 11. Trends Microbiol. 2020. PMID: 32536523 Free PMC article. Review.
-
Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication.PLoS Pathog. 2012;8(12):e1003056. doi: 10.1371/journal.ppat.1003056. Epub 2012 Dec 6. PLoS Pathog. 2012. PMID: 23236278 Free PMC article.
-
Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes.J Gen Virol. 2010 Sep;91(Pt 9):2230-7. doi: 10.1099/vir.0.022186-0. Epub 2010 May 19. J Gen Virol. 2010. PMID: 20484561
-
Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment.J Virol. 2013 Oct;87(19):10612-27. doi: 10.1128/JVI.01370-13. Epub 2013 Jul 24. J Virol. 2013. PMID: 23885072 Free PMC article.
-
The double-membrane vesicle (DMV): a virus-induced organelle dedicated to the replication of SARS-CoV-2 and other positive-sense single-stranded RNA viruses.Cell Mol Life Sci. 2022 Jul 16;79(8):425. doi: 10.1007/s00018-022-04469-x. Cell Mol Life Sci. 2022. PMID: 35841484 Free PMC article. Review.
Cited by
-
Biogenesis and architecture of arterivirus replication organelles.Virus Res. 2016 Jul 15;220:70-90. doi: 10.1016/j.virusres.2016.04.001. Epub 2016 Apr 9. Virus Res. 2016. PMID: 27071852 Free PMC article. Review.
-
What role for cellular metabolism in the control of hepatitis viruses?Front Immunol. 2022 Nov 17;13:1033314. doi: 10.3389/fimmu.2022.1033314. eCollection 2022. Front Immunol. 2022. PMID: 36466918 Free PMC article. Review.
-
Visualisation and analysis of hepatitis C virus non-structural proteins using super-resolution microscopy.Sci Rep. 2018 Sep 11;8(1):13604. doi: 10.1038/s41598-018-31861-0. Sci Rep. 2018. PMID: 30206266 Free PMC article.
-
Regulatory Role of Phospholipids in Hepatitis C Virus Replication and Protein Function.Pathogens. 2022 Jan 15;11(1):102. doi: 10.3390/pathogens11010102. Pathogens. 2022. PMID: 35056049 Free PMC article. Review.
-
Double-Membrane Vesicles as Platforms for Viral Replication.Trends Microbiol. 2020 Dec;28(12):1022-1033. doi: 10.1016/j.tim.2020.05.009. Epub 2020 Jun 11. Trends Microbiol. 2020. PMID: 32536523 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

