Transposable elements reveal a stem cell-specific class of long noncoding RNAs
- PMID: 23181609
- PMCID: PMC3580499
- DOI: 10.1186/gb-2012-13-11-r107
Transposable elements reveal a stem cell-specific class of long noncoding RNAs
Abstract
Background: Numerous studies over the past decade have elucidated a large set of long intergenic noncoding RNAs (lincRNAs) in the human genome. Research since has shown that lincRNAs constitute an important layer of genome regulation across a wide spectrum of species. However, the factors governing their evolution and origins remain relatively unexplored. One possible factor driving lincRNA evolution and biological function is transposable element (TE) insertions. Here, we comprehensively characterize the TE content of lincRNAs relative to genomic averages and protein coding transcripts.
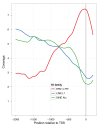
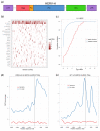
Results: Our analysis of the TE composition of 9,241 human lincRNAs revealed that, in sharp contrast to protein coding genes, 83% of lincRNAs contain a TE, and TEs comprise 42% of lincRNA sequence. lincRNA TE composition varies significantly from genomic averages - L1 and Alu elements are depleted and broad classes of endogenous retroviruses are enriched. TEs occur in biased positions and orientations within lincRNAs, particularly at their transcription start sites, suggesting a role in lincRNA transcriptional regulation. Accordingly, we observed a dramatic example of HERVH transcriptional regulatory signals correlating strongly with stem cell-specific expression of lincRNAs. Conversely, lincRNAs devoid of TEs are expressed at greater levels than lincRNAs with TEs in all tissues and cell lines, particularly in the testis.
Conclusions: TEs pervade lincRNAs, dividing them into classes, and may have shaped lincRNA evolution and function by conferring tissue-specific expression from extant transcriptional regulatory signals.
Figures



Similar articles
-
Comparative analysis of lincRNA in insect species.BMC Evol Biol. 2017 Jul 3;17(1):155. doi: 10.1186/s12862-017-0985-0. BMC Evol Biol. 2017. PMID: 28673235 Free PMC article.
-
Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants.Plant J. 2017 Apr;90(1):133-146. doi: 10.1111/tpj.13481. Plant J. 2017. PMID: 28106309 Free PMC article.
-
Comparative genomic analyses highlight the contribution of pseudogenized protein-coding genes to human lincRNAs.BMC Genomics. 2017 Oct 16;18(1):786. doi: 10.1186/s12864-017-4156-x. BMC Genomics. 2017. PMID: 29037146 Free PMC article.
-
The intertwining of transposable elements and non-coding RNAs.Int J Mol Sci. 2013 Jun 26;14(7):13307-28. doi: 10.3390/ijms140713307. Int J Mol Sci. 2013. PMID: 23803660 Free PMC article. Review.
-
How to tame an endogenous retrovirus: HERVH and the evolution of human pluripotency.Curr Opin Virol. 2017 Aug;25:49-58. doi: 10.1016/j.coviro.2017.07.001. Epub 2017 Jul 24. Curr Opin Virol. 2017. PMID: 28750248 Review.
Cited by
-
Hidden magicians of genome evolution.Indian J Med Res. 2013 Jun;137(6):1052-60. Indian J Med Res. 2013. PMID: 23852286 Free PMC article. Review.
-
Transposable elements in the mammalian embryo: pioneers surviving through stealth and service.Genome Biol. 2016 May 9;17:100. doi: 10.1186/s13059-016-0965-5. Genome Biol. 2016. PMID: 27161170 Free PMC article. Review.
-
Evolution to the rescue: using comparative genomics to understand long non-coding RNAs.Nat Rev Genet. 2016 Oct;17(10):601-14. doi: 10.1038/nrg.2016.85. Epub 2016 Aug 30. Nat Rev Genet. 2016. PMID: 27573374 Review.
-
A call for benchmarking transposable element annotation methods.Mob DNA. 2015 Aug 4;6:13. doi: 10.1186/s13100-015-0044-6. eCollection 2015. Mob DNA. 2015. PMID: 26244060 Free PMC article.
-
The transcriptional trajectories of pluripotency and differentiation comprise genes with antithetical architecture and repetitive-element content.BMC Biol. 2021 Mar 25;19(1):60. doi: 10.1186/s12915-020-00928-8. BMC Biol. 2021. PMID: 33765992 Free PMC article.
References
-
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

