STAT1:DNA sequence-dependent binding modulation by phosphorylation, protein:protein interactions and small-molecule inhibition
- PMID: 23180800
- PMCID: PMC3553987
- DOI: 10.1093/nar/gks1085
STAT1:DNA sequence-dependent binding modulation by phosphorylation, protein:protein interactions and small-molecule inhibition
Abstract
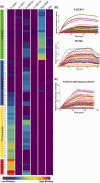
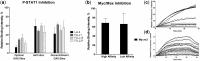
The DNA-binding specificity and affinity of the dimeric human transcription factor (TF) STAT1, were assessed by total internal reflectance fluorescence protein-binding microarrays (TIRF-PBM) to evaluate the effects of protein phosphorylation, higher-order polymerization and small-molecule inhibition. Active, phosphorylated STAT1 showed binding preferences consistent with prior characterization, whereas unphosphorylated STAT1 showed a weak-binding preference for one-half of the GAS consensus site, consistent with recent models of STAT1 structure and function in response to phosphorylation. This altered-binding preference was further tested by use of the inhibitor LLL3, which we show to disrupt STAT1 binding in a sequence-dependent fashion. To determine if this sequence-dependence is specific to STAT1 and not a general feature of human TF biology, the TF Myc/Max was analysed and tested with the inhibitor Mycro3. Myc/Max inhibition by Mycro3 is sequence independent, suggesting that the sequence-dependent inhibition of STAT1 may be specific to this system and a useful target for future inhibitor design.
Figures
Similar articles
-
Biophysical characterization of the b-HLH-LZ of ΔMax, an alternatively spliced isoform of Max found in tumor cells: Towards the validation of a tumor suppressor role for the Max homodimers.PLoS One. 2017 Mar 28;12(3):e0174413. doi: 10.1371/journal.pone.0174413. eCollection 2017. PLoS One. 2017. PMID: 28350847 Free PMC article.
-
Myc phosphorylation in its basic helix-loop-helix region destabilizes transient α-helical structures, disrupting Max and DNA binding.J Biol Chem. 2018 Jun 15;293(24):9301-9310. doi: 10.1074/jbc.RA118.002709. Epub 2018 Apr 25. J Biol Chem. 2018. PMID: 29695509 Free PMC article.
-
Sequence-specific DNA binding by MYC/MAX to low-affinity non-E-box motifs.PLoS One. 2017 Jul 18;12(7):e0180147. doi: 10.1371/journal.pone.0180147. eCollection 2017. PLoS One. 2017. PMID: 28719624 Free PMC article.
-
Structural aspects of interactions within the Myc/Max/Mad network.Curr Top Microbiol Immunol. 2006;302:123-43. doi: 10.1007/3-540-32952-8_5. Curr Top Microbiol Immunol. 2006. PMID: 16620027 Review.
-
Small-molecule modulators of c-Myc/Max and Max/Max interactions.Curr Top Microbiol Immunol. 2011;348:139-49. doi: 10.1007/82_2010_90. Curr Top Microbiol Immunol. 2011. PMID: 20680803 Review.
Cited by
-
Quantitative specificity of STAT1 and several variants.Nucleic Acids Res. 2017 Aug 21;45(14):8199-8207. doi: 10.1093/nar/gkx393. Nucleic Acids Res. 2017. PMID: 28510715 Free PMC article.
-
Targeting RNA polymerase I to treat MYC-driven cancer.Oncogene. 2015 Jan 22;34(4):403-12. doi: 10.1038/onc.2014.13. Epub 2014 Mar 10. Oncogene. 2015. PMID: 24608428 Review.
-
Using protein-binding microarrays to study transcription factor specificity: homologs, isoforms and complexes.Brief Funct Genomics. 2015 Jan;14(1):17-29. doi: 10.1093/bfgp/elu046. Epub 2014 Nov 26. Brief Funct Genomics. 2015. PMID: 25431149 Free PMC article. Review.
-
Inhibition of complement C3 prevents osteoarthritis progression in guinea pigs by blocking STAT1 activation.Commun Biol. 2024 Mar 27;7(1):370. doi: 10.1038/s42003-024-06051-6. Commun Biol. 2024. PMID: 38538870 Free PMC article.
-
Dry and wet approaches for genome-wide functional annotation of conventional and unconventional transcriptional activators.Comput Struct Biotechnol J. 2016 Jun 29;14:262-70. doi: 10.1016/j.csbj.2016.06.004. eCollection 2016. Comput Struct Biotechnol J. 2016. PMID: 27453771 Free PMC article. Review.
References
-
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. - PubMed
-
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. - PubMed
-
- Levy DE, Darnell JE. STATs: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. - PubMed
-
- Horak CE, Snyder M. ChIP-chip: a genomic approach for identifying transcription factor binding sites. Guide Yeast Genet. Mol. Cell Biol. 2002 Pt B, 350, 469–483. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

