An S/T-Q cluster domain census unveils new putative targets under Tel1/Mec1 control
- PMID: 23176708
- PMCID: PMC3564818
- DOI: 10.1186/1471-2164-13-664
An S/T-Q cluster domain census unveils new putative targets under Tel1/Mec1 control
Abstract
Background: The cellular response to DNA damage is immediate and highly coordinated in order to maintain genome integrity and proper cell division. During the DNA damage response (DDR), the sensor kinases Tel1 and Mec1 in Saccharomyces cerevisiae and ATM and ATR in human, phosphorylate multiple mediators which activate effector proteins to initiate cell cycle checkpoints and DNA repair. A subset of kinase substrates are recognized by the S/T-Q cluster domain (SCD), which contains motifs of serine (S) or threonine (T) followed by a glutamine (Q). However, the full repertoire of proteins and pathways controlled by Tel1 and Mec1 is unknown.
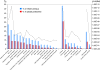
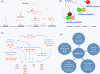
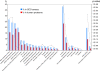
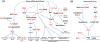
Results: To identify all putative SCD-containing proteins, we analyzed the distribution of S/T-Q motifs within verified Tel1/Mec1 targets and arrived at a unifying SCD definition of at least 3 S/T-Q within a stretch of 50 residues. This new SCD definition was used in a custom bioinformatics pipeline to generate a census of SCD-containing proteins in both yeast and human. In yeast, 436 proteins were identified, a significantly larger number of hits than were expected by chance. These SCD-containing proteins did not distribute equally across GO-ontology terms, but were significantly enriched for those involved in processes related to the DDR. We also found a significant enrichment of proteins involved in telophase and cytokinesis, protein transport and endocytosis suggesting possible novel Tel1/Mec1 targets in these pathways. In the human proteome, a wide range of similar proteins were identified, including homologs of some SCD-containing proteins found in yeast. This list also included high concentrations of proteins in the Mediator, spindle pole body/centrosome and actin cytoskeleton complexes.
Conclusions: Using a bioinformatic approach, we have generated a census of SCD-containing proteins that are involved not only in known DDR pathways but several other pathways under Tel1/Mec1 control suggesting new putative targets for these kinases.
Figures


Similar articles
-
Dominant TEL1-hy mutations compensate for Mec1 lack of functions in the DNA damage response.Mol Cell Biol. 2008 Jan;28(1):358-75. doi: 10.1128/MCB.01214-07. Epub 2007 Oct 22. Mol Cell Biol. 2008. PMID: 17954565 Free PMC article.
-
Budding yeast Rad51: a paradigm for how phosphorylation and intrinsic structural disorder regulate homologous recombination and protein homeostasis.Curr Genet. 2021 Jun;67(3):389-396. doi: 10.1007/s00294-020-01151-2. Epub 2021 Jan 12. Curr Genet. 2021. PMID: 33433732 Free PMC article. Review.
-
Tel1 Activation by the MRX Complex Is Sufficient for Telomere Length Regulation but Not for the DNA Damage Response in Saccharomyces cerevisiae.Genetics. 2019 Dec;213(4):1271-1288. doi: 10.1534/genetics.119.302713. Epub 2019 Oct 23. Genetics. 2019. PMID: 31645360 Free PMC article.
-
Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination.Cell. 2008 Mar 7;132(5):758-70. doi: 10.1016/j.cell.2008.01.035. Cell. 2008. PMID: 18329363
-
Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks.DNA Repair (Amst). 2013 Oct;12(10):791-9. doi: 10.1016/j.dnarep.2013.07.009. Epub 2013 Aug 13. DNA Repair (Amst). 2013. PMID: 23953933 Review.
Cited by
-
DNA damage response and spindle assembly checkpoint function throughout the cell cycle to ensure genomic integrity.PLoS Genet. 2015 Apr 21;11(4):e1005150. doi: 10.1371/journal.pgen.1005150. eCollection 2015 Apr. PLoS Genet. 2015. PMID: 25898113 Free PMC article.
-
The leukemia-associated Rho guanine nucleotide exchange factor LARG is required for efficient replication stress signaling.Cell Cycle. 2014;13(21):3450-9. doi: 10.4161/15384101.2014.956529. Cell Cycle. 2014. PMID: 25485589 Free PMC article.
-
The spindle assembly checkpoint: More than just keeping track of the spindle.Trends Cell Mol Biol. 2015;10:141-150. Trends Cell Mol Biol. 2015. PMID: 27667906 Free PMC article.
-
Cross-Talk between Carbon Metabolism and the DNA Damage Response in S. cerevisiae.Cell Rep. 2015 Sep 22;12(11):1865-75. doi: 10.1016/j.celrep.2015.08.025. Epub 2015 Sep 3. Cell Rep. 2015. PMID: 26344768 Free PMC article.
-
Noncanonical usage of stop codons in ciliates expands proteins with structurally flexible Q-rich motifs.Elife. 2024 Feb 23;12:RP91405. doi: 10.7554/eLife.91405. Elife. 2024. PMID: 38393970 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

