Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile
- PMID: 23175653
- PMCID: PMC3554014
- DOI: 10.1128/JB.01980-12
Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile
Abstract
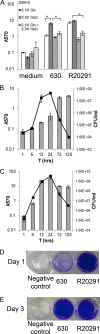
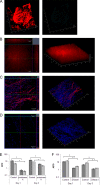
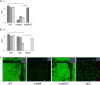
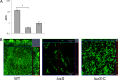
Bacteria within biofilms are protected from multiple stresses, including immune responses and antimicrobial agents. The biofilm-forming ability of bacterial pathogens has been associated with increased antibiotic resistance and chronic recurrent infections. Although biofilms have been well studied for several gut pathogens, little is known about biofilm formation by anaerobic gut species. The obligate anaerobe Clostridium difficile causes C. difficile infection (CDI), a major health care-associated problem primarily due to the high incidence of recurring infections. C. difficile colonizes the gut when the normal intestinal microflora is disrupted by antimicrobial agents; however, the factors or processes involved in gut colonization during infection remain unclear. We demonstrate that clinical C. difficile strains, i.e., strain 630 and the hypervirulent strain R20291, form structured biofilms in vitro, with R20291 accumulating substantially more biofilm. Microscopic and biochemical analyses show multiple layers of bacteria encased in a biofilm matrix containing proteins, DNA, and polysaccharide. Employing isogenic mutants, we show that virulence-associated proteins, Cwp84, flagella, and a putative quorum-sensing regulator, LuxS, are all required for maximal biofilm formation by C. difficile. Interestingly, a mutant in Spo0A, a transcription factor that controls spore formation, was defective for biofilm formation, indicating a possible link between sporulation and biofilm formation. Furthermore, we demonstrate that bacteria in clostridial biofilms are more resistant to high concentrations of vancomycin, a drug commonly used for treatment of CDI. Our data suggest that biofilm formation by C. difficile is a complex multifactorial process and may be a crucial mechanism for clostridial persistence in the host.
Figures

Comment in
-
Biofilm formation by Clostridium difficile.Gut Microbes. 2013 Sep-Oct;4(5):397-402. doi: 10.4161/gmic.25862. Epub 2013 Jul 25. Gut Microbes. 2013. PMID: 23892245 Free PMC article.
Similar articles
-
High sporulation and overexpression of virulence factors in biofilms and reduced susceptibility to vancomycin and linezolid in recurrent Clostridium [Clostridioides] difficile infection isolates.PLoS One. 2019 Jul 31;14(7):e0220671. doi: 10.1371/journal.pone.0220671. eCollection 2019. PLoS One. 2019. PMID: 31365590 Free PMC article.
-
Biofilm formation by Clostridium difficile.Gut Microbes. 2013 Sep-Oct;4(5):397-402. doi: 10.4161/gmic.25862. Epub 2013 Jul 25. Gut Microbes. 2013. PMID: 23892245 Free PMC article.
-
Biofilm regulation in Clostridioides difficile: Novel systems linked to hypervirulence.PLoS Pathog. 2021 Sep 9;17(9):e1009817. doi: 10.1371/journal.ppat.1009817. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34499698 Free PMC article. Review.
-
The role of single and mixed biofilms in Clostridioides difficile infection and strategies for prevention and inhibition.Crit Rev Microbiol. 2024 May;50(3):285-299. doi: 10.1080/1040841X.2023.2189950. Epub 2023 Mar 20. Crit Rev Microbiol. 2024. PMID: 36939635 Review.
-
Characterisation of Clostridium difficile biofilm formation, a role for Spo0A.PLoS One. 2012;7(12):e50527. doi: 10.1371/journal.pone.0050527. Epub 2012 Dec 7. PLoS One. 2012. PMID: 23236376 Free PMC article.
Cited by
-
Does the Gut Microbiota Modulate Host Physiology through Polymicrobial Biofilms?Microbes Environ. 2020;35(3):ME20037. doi: 10.1264/jsme2.ME20037. Microbes Environ. 2020. PMID: 32624527 Free PMC article. Review.
-
Characterisation of Clostridium difficile strains isolated from Groote Schuur Hospital, Cape Town, South Africa.Eur J Clin Microbiol Infect Dis. 2016 Oct;35(10):1709-18. doi: 10.1007/s10096-016-2717-6. Epub 2016 Jul 27. Eur J Clin Microbiol Infect Dis. 2016. PMID: 27465145
-
Clostridioides difficile Flagella.Int J Mol Sci. 2024 Feb 12;25(4):2202. doi: 10.3390/ijms25042202. Int J Mol Sci. 2024. PMID: 38396876 Free PMC article. Review.
-
Overcoming Antibiotic Resistance with Novel Paradigms of Antibiotic Selection.Microorganisms. 2022 Nov 30;10(12):2383. doi: 10.3390/microorganisms10122383. Microorganisms. 2022. PMID: 36557636 Free PMC article. Review.
-
Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing.Nat Commun. 2019 Oct 4;10(1):4544. doi: 10.1038/s41467-019-12448-3. Nat Commun. 2019. PMID: 31586051 Free PMC article.
References
-
- Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends. Microbiol. 13:20–26 - PubMed
-
- Cummings JH, Antoine JM, Azpiroz F, Bourdet-Sicard R, Brandtzaeg P, Calder PC, Gibson GR, Guarner F, Isolauri E, Pannemans D, Shortt C, Tuijtelaars S, Watzl B. 2004. PASSCLAIM: gut health and immunity. Eur. J. Nutr. 43(Suppl 2):II118–II173 - PubMed
-
- Macfarlane S, Dillon JF. 2007. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 102:1187–1196 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

