Histone acetyltransferases: Rising ancient counterparts to protein kinases
- PMID: 23175385
- PMCID: PMC4017165
- DOI: 10.1002/bip.22128
Histone acetyltransferases: Rising ancient counterparts to protein kinases
Abstract
Protein kinases catalyze phosphorylation, a posttranslational modification widely utilized in cell signaling. Histone acetyltransferases (HATs) catalyze a counterpart posttranslational modification of acetylation which marks histones for epigenetic signaling but in some cases modifies non-histone proteins to mediate other cellular activities. In addition, recent proteomic studies have revealed that thousands of proteins are acetylated throughout the cell to regulate diverse biological processes, thus placing acetyltransferases on the same playing field as kinases. Emerging biochemical and structural data further supports mechanistic and biological links between the two enzyme families. In this article, we will review what is known to date about the structure, catalysis and mode of regulation of HAT enzymes and draw analogies, where relevant, to protein kinases. This comparison reveals that HATs may be rising ancient counterparts to protein kinases.
Copyright © 2012 Wiley Periodicals, Inc.
Figures
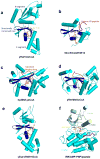
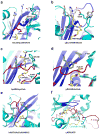
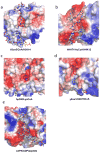
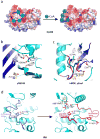
Similar articles
-
Structure and mechanism of non-histone protein acetyltransferase enzymes.FEBS J. 2013 Nov;280(22):5570-81. doi: 10.1111/febs.12373. Epub 2013 Jun 28. FEBS J. 2013. PMID: 23742047 Free PMC article. Review.
-
Catalysis and substrate selection by histone/protein lysine acetyltransferases.Curr Opin Struct Biol. 2008 Dec;18(6):682-9. doi: 10.1016/j.sbi.2008.11.004. Curr Opin Struct Biol. 2008. PMID: 19056256 Free PMC article. Review.
-
Writers and readers of histone acetylation: structure, mechanism, and inhibition.Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a018762. doi: 10.1101/cshperspect.a018762. Cold Spring Harb Perspect Biol. 2014. PMID: 24984779 Free PMC article. Review.
-
Histone acetyltransferases: challenges in targeting bi-substrate enzymes.Clin Epigenetics. 2016 May 26;8:59. doi: 10.1186/s13148-016-0225-2. eCollection 2016. Clin Epigenetics. 2016. PMID: 27231488 Free PMC article. Review.
-
The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.Nat Struct Biol. 2002 Nov;9(11):862-9. doi: 10.1038/nsb849. Nat Struct Biol. 2002. PMID: 12368900
Cited by
-
Acetyl-CoA: An interplay between metabolism and epigenetics in cancer.Front Mol Med. 2022 Nov 16;2:1044585. doi: 10.3389/fmmed.2022.1044585. eCollection 2022. Front Mol Med. 2022. PMID: 39086974 Free PMC article. Review.
-
The molecular basis for histone H4- and H2A-specific amino-terminal acetylation by NatD.Structure. 2015 Feb 3;23(2):332-41. doi: 10.1016/j.str.2014.10.025. Epub 2015 Jan 22. Structure. 2015. PMID: 25619998 Free PMC article.
-
Synchronous recruitment of epigenetic modifiers to endotoxin synergistically activated Tnf-α gene in acute kidney injury.PLoS One. 2013 Jul 30;8(7):e70322. doi: 10.1371/journal.pone.0070322. Print 2013. PLoS One. 2013. PMID: 23936185 Free PMC article.
-
Impact of Epigenetic Dietary Components on Cancer through Histone Modifications.Curr Med Chem. 2015;22(17):2051-64. doi: 10.2174/0929867322666150420102641. Curr Med Chem. 2015. PMID: 25891109 Free PMC article. Review.
-
An evolving understanding of nuclear receptor coregulator proteins.J Mol Endocrinol. 2013 Nov 7;51(3):T23-36. doi: 10.1530/JME-13-0227. Print 2013 Dec. J Mol Endocrinol. 2013. PMID: 24203923 Free PMC article. Review.
References
-
- Krebs EG, Fischer EH. J Biol Chem. 1955;216:113–120. - PubMed
-
- Fischer EH, Krebs EG. J Biol Chem. 1955;216:121–132. - PubMed
-
- Walsh DA, Perkins JP, Krebs EG. Journal of Biological Chemistry. 1968;243:3763–3765. - PubMed
-
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Cell. 1996;84:843–851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

