Gene knockout and knockin by zinc-finger nucleases: current status and perspectives
- PMID: 23161061
- PMCID: PMC11113862
- DOI: 10.1007/s00018-012-1204-1
Gene knockout and knockin by zinc-finger nucleases: current status and perspectives
Abstract
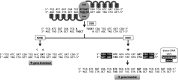
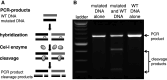
Zinc-finger nucleases (ZFNs) are engineered site-specific DNA cleavage enzymes that may be designed to recognize long target sites and thus cut DNA with high specificity. ZFNs mediate permanent and targeted genetic alteration via induction of a double-strand break at a specific genomic site. Compared to conventional homology-based gene targeting, ZFNs can increase the targeting rate by up to 100,000-fold; gene disruption via mutagenic DNA repair is similarly efficient. The utility of ZFNs has been shown in many organisms, including insects, amphibians, plants, nematodes, and several mammals, including humans. This broad range of tractable species renders ZFNs a useful tool for improving the understanding of complex physiological systems, to produce transgenic animals, cell lines, and plants, and to treat human disease.
Figures



Similar articles
-
Advances in genetic modification of farm animals using zinc-finger nucleases (ZFN).Chromosome Res. 2015 Feb;23(1):7-15. doi: 10.1007/s10577-014-9451-7. Chromosome Res. 2015. PMID: 25596823 Review.
-
Zinc-finger nucleases: a powerful tool for genetic engineering of animals.Transgenic Res. 2010 Jun;19(3):363-71. doi: 10.1007/s11248-009-9323-7. Epub 2009 Sep 26. Transgenic Res. 2010. PMID: 19821047 Review.
-
Basics of genome editing technology and its application in livestock species.Reprod Domest Anim. 2017 Aug;52 Suppl 3:4-13. doi: 10.1111/rda.13012. Reprod Domest Anim. 2017. PMID: 28815851 Review.
-
Genome editing with CompoZr custom zinc finger nucleases (ZFNs).J Vis Exp. 2012 Jun 14;(64):e3304. doi: 10.3791/3304. J Vis Exp. 2012. PMID: 22732945 Free PMC article.
-
ZFNGenome: a comprehensive resource for locating zinc finger nuclease target sites in model organisms.BMC Genomics. 2011 Jan 28;12:83. doi: 10.1186/1471-2164-12-83. BMC Genomics. 2011. PMID: 21276248 Free PMC article.
Cited by
-
Advances in genetic modification of farm animals using zinc-finger nucleases (ZFN).Chromosome Res. 2015 Feb;23(1):7-15. doi: 10.1007/s10577-014-9451-7. Chromosome Res. 2015. PMID: 25596823 Review.
-
Programmable Molecular Scissors: Applications of a New Tool for Genome Editing in Biotech.Mol Ther Nucleic Acids. 2019 Mar 1;14:212-238. doi: 10.1016/j.omtn.2018.11.016. Epub 2018 Dec 6. Mol Ther Nucleic Acids. 2019. PMID: 30641475 Free PMC article. Review.
-
Genomic Knockout of Endogenous Canine P-Glycoprotein in Wild-Type, Human P-Glycoprotein and Human BCRP Transfected MDCKII Cell Lines by Zinc Finger Nucleases.Pharm Res. 2015 Jun;32(6):2060-71. doi: 10.1007/s11095-014-1599-5. Epub 2014 Dec 19. Pharm Res. 2015. PMID: 25522789
-
Gene-edited cells: novel allogeneic gene/cell therapy for epidermolysis bullosa.J Appl Genet. 2024 Dec;65(4):705-726. doi: 10.1007/s13353-024-00839-2. Epub 2024 Mar 9. J Appl Genet. 2024. PMID: 38459407 Review.
-
Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family.Mob DNA. 2015 Oct 2;6:17. doi: 10.1186/s13100-015-0050-8. eCollection 2015. Mob DNA. 2015. PMID: 26435754 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

