Sphingomyelin synthase 1 activity is regulated by the BCR-ABL oncogene
- PMID: 23160178
- PMCID: PMC3617953
- DOI: 10.1194/jlr.M033985
Sphingomyelin synthase 1 activity is regulated by the BCR-ABL oncogene
Abstract
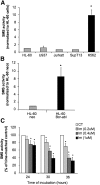
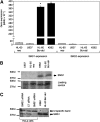
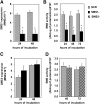
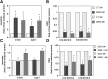
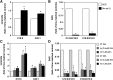
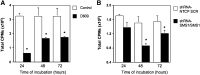
Sphingomyelin synthase (SMS) produces sphingomyelin while consuming ceramide (a negative regulator of cell proliferation) and forming diacylglycerol (DAG) (a mitogenic factor). Therefore, enhanced SMS activity could favor cell proliferation. To examine if dysregulated SMS contributes to leukemogenesis, we measured SMS activity in several leukemic cell lines and found that it is highly elevated in K562 chronic myelogenous leukemia (CML) cells. The increased SMS in K562 cells was caused by the presence of Bcr-abl, a hallmark of CML; stable expression of Bcr-abl elevated SMS activity in HL-60 cells while inhibition of the tyrosine kinase activity of Bcr-abl with Imatinib mesylate decreased SMS activity in K562 cells. The increased SMS activity was the result of up-regulation of the Sms1 isoform. Inhibition of SMS activity with D609 (a pharmacological SMS inhibitor) or down-regulation of SMS1 expression by siRNA selectively inhibited the proliferation of Bcr-abl-positive cells. The inhibition was associated with an increased production of ceramide and a decreased production of DAG, conditions that antagonize cell proliferation. A similar change in lipid profile was also observed upon pharmacological inhibition of Bcr-abl (K526 cells) and siRNA-mediated down-regulation of BCR-ABL (HL-60/Bcr-abl cells). These findings indicate that Sms1 is a downstream target of Bcr-abl, involved in sustaining cell proliferation of Bcr-abl-positive cells.
Figures

Similar articles
-
Bcr-Abl regulation of sphingomyelin synthase 1 reveals a novel oncogenic-driven mechanism of protein up-regulation.FASEB J. 2018 Aug;32(8):4270-4283. doi: 10.1096/fj.201701016R. Epub 2018 Mar 13. FASEB J. 2018. PMID: 29533737 Free PMC article.
-
Inhibition of sphingomyelin synthase (SMS) affects intracellular sphingomyelin accumulation and plasma membrane lipid organization.Biochim Biophys Acta. 2007 Sep;1771(9):1186-94. doi: 10.1016/j.bbalip.2007.05.007. Epub 2007 Jun 6. Biochim Biophys Acta. 2007. PMID: 17616479 Free PMC article.
-
Sphingomyelin synthase, a potential regulator of intracellular levels of ceramide and diacylglycerol during SV40 transformation. Does sphingomyelin synthase account for the putative phosphatidylcholine-specific phospholipase C?J Biol Chem. 1998 Jun 5;273(23):14550-9. doi: 10.1074/jbc.273.23.14550. J Biol Chem. 1998. PMID: 9603970
-
Mechanisms of resistance to imatinib mesylate in Bcr-Abl-positive leukemias.Curr Opin Oncol. 2002 Nov;14(6):616-20. doi: 10.1097/00001622-200211000-00005. Curr Opin Oncol. 2002. PMID: 12409651 Review.
-
Chronic myelogenous leukemia: from molecular biology to clinical aspects and novel targeted therapies.J Nippon Med Sch. 2006 Aug;73(4):178-92. doi: 10.1272/jnms.73.178. J Nippon Med Sch. 2006. PMID: 16936444 Review.
Cited by
-
Phosphorylation of serine palmitoyltransferase long chain-1 (SPTLC1) on tyrosine 164 inhibits its activity and promotes cell survival.J Biol Chem. 2013 Jun 14;288(24):17190-201. doi: 10.1074/jbc.M112.409185. Epub 2013 Apr 29. J Biol Chem. 2013. PMID: 23629659 Free PMC article.
-
Ceramide Transfer Protein (CERT): An Overlooked Molecular Player in Cancer.Int J Mol Sci. 2021 Dec 7;22(24):13184. doi: 10.3390/ijms222413184. Int J Mol Sci. 2021. PMID: 34947980 Free PMC article. Review.
-
Regulation of human sphingomyelin synthase 1 translation through its 5'-untranslated region.FEBS Lett. 2020 Nov;594(22):3751-3764. doi: 10.1002/1873-3468.13952. Epub 2020 Oct 31. FEBS Lett. 2020. PMID: 33037626 Free PMC article.
-
Bcr-Abl regulation of sphingomyelin synthase 1 reveals a novel oncogenic-driven mechanism of protein up-regulation.FASEB J. 2018 Aug;32(8):4270-4283. doi: 10.1096/fj.201701016R. Epub 2018 Mar 13. FASEB J. 2018. PMID: 29533737 Free PMC article.
-
Cholesterol and Sphingolipid Enriched Lipid Rafts as Therapeutic Targets in Cancer.Int J Mol Sci. 2021 Jan 13;22(2):726. doi: 10.3390/ijms22020726. Int J Mol Sci. 2021. PMID: 33450869 Free PMC article. Review.
References
-
- Bernert J. T., Ullman M. D. 1981. Biosynthesis of sphingomyelin from erythro-ceramides and phosphatidylcholine by a microsomal cholinephosphoesterase. Biochim. Biophys. Acta. 666: 99–109 - PubMed
-
- Hatch G. M., Vance D. E. 1992. Stimulation of sphingomyelin biosynthesis by brefeldin A and sphingomyelin breakdown by okadaic acid treatment of rat hepatocytes. J. Biol. Chem. 267: 12443–12451 - PubMed
-
- Marggraf W. D., Anderer F. A., Kanfer J. 1981. The formation of sphingomyelin from phosphatidylcholine in plasma membrane preparations from mouse fibroblasts. Biochim. Biophys. Acta. 664: 61–73 - PubMed
-
- Marggraf W. D., Zertani R., Anderer F. A., Kanfer J. N. 1982. The role of endogenous phosphatidylcholine and ceramide in the biosynthesis of sphingomyelin in mouse fibroblasts. Biochim. Biophys. Acta. 710: 314–323 - PubMed
-
- Merrill A. H., Jr, Jones D. D. 1990. An update of the enzymology and regulation of sphingomyelin metabolism. Biochim.Biophys.Acta Lipids Lipid Metab. 1044: 1–12 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

