Characterization of a chitin synthase encoding gene and effect of diflubenzuron in soybean aphid, Aphis glycines
- PMID: 23139631
- PMCID: PMC3492791
- DOI: 10.7150/ijbs.4189
Characterization of a chitin synthase encoding gene and effect of diflubenzuron in soybean aphid, Aphis glycines
Abstract
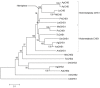
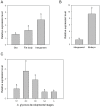
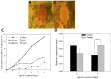
Chitin synthases are critical enzymes for synthesis of chitin and thus for subsequent growth and development in insects. We identified the cDNA of chitin synthase gene (CHS) in Aphis glycines, the soybean aphid, which is a serious pest of soybean. The full-length cDNA of CHS in A. glycines (AyCHS) was 5802 bp long with an open reading frame of 4704 bp that encoded for a 1567 amino acid residues protein. The predicted AyCHS protein had a molecular mass of 180.05 kDa and its amino acid sequence contained all the signature motifs (EDR, QRRRW and TWGTR) of chitin synthases. The quantitative real-time PCR (qPCR) analysis revealed that AyCHS was expressed in all major tissues (gut, fat body and integument); however, it had the highest expression in integument (~3.5 fold compared to gut). Interestingly, the expression of AyCHS in developing embryos was nearly 7 fold higher compared to adult integument, which probably is a reflection of embryonic molts in hemimetabolus insects. Expression analysis in different developmental stages of A. glycines revealed a consistent AyCHS expression in all stages. Further, through leaf dip bioassay, we tested the effect of diflubenzuron (DFB, Dimilin ®), a chitin-synthesis inhibitor, on A. glycines' survival, fecundity and body weight. When fed with soybean leaves previously dipped in 50 ppm DFB solution, A. glycines nymphs suffered significantly higher mortality compared to control. A. glycines nymphs feeding on diflubenzuron treated leaves showed a slightly enhanced expression (1.67 fold) of AyCHS compared to nymphs on untreated leaves. We discussed the potential applications of the current study to develop novel management strategies using chitin-synthesis inhibitors and using RNAi by knocking down AyCHS expression.
Keywords: Aphis glycines; Chitin synthase; Diflubenzuron.; Embryo; Integument.
Conflict of interest statement
Competing Interests: The author(s) declare that they have no competing interests.
Figures



Similar articles
-
Identification, characterization and functional analysis of a chitin synthase gene in the brown citrus aphid, Toxoptera citricida (Hemiptera, Aphididae).Insect Mol Biol. 2016 Aug;25(4):422-30. doi: 10.1111/imb.12228. Epub 2016 Mar 17. Insect Mol Biol. 2016. PMID: 26991909
-
Silencing of the Chitin Synthase Gene Is Lethal to the Asian Citrus Psyllid, Diaphorina citri.Int J Mol Sci. 2019 Jul 31;20(15):3734. doi: 10.3390/ijms20153734. Int J Mol Sci. 2019. PMID: 31370145 Free PMC article.
-
Molecular characterization and expression analysis of soluble trehalase gene in Aphis glycines, a migratory pest of soybean.Bull Entomol Res. 2013 Jun;103(3):286-95. doi: 10.1017/S0007485312000697. Epub 2013 Feb 28. Bull Entomol Res. 2013. PMID: 23445549
-
Characterization of a chitin synthase cDNA and its increased mRNA level associated with decreased chitin synthesis in Anopheles quadrimaculatus exposed to diflubenzuron.Insect Biochem Mol Biol. 2006 Sep;36(9):712-25. doi: 10.1016/j.ibmb.2006.06.002. Epub 2006 Jun 16. Insect Biochem Mol Biol. 2006. PMID: 16935220
-
The Resistant Soybean-Aphis glycines Interaction: Current Knowledge and Prospects.Front Plant Sci. 2020 Aug 11;11:1223. doi: 10.3389/fpls.2020.01223. eCollection 2020. Front Plant Sci. 2020. PMID: 32849757 Free PMC article. Review.
Cited by
-
Effects of RNAi-based silencing of chitin synthase gene on moulting and fecundity in pea aphids (Acyrthosiphon pisum).Sci Rep. 2019 Mar 6;9(1):3694. doi: 10.1038/s41598-019-39837-4. Sci Rep. 2019. PMID: 30842508 Free PMC article.
-
Uninterrupted Development of Two Aphid Species Belonging to Cinara Genus during Winter Diapause.Insects. 2020 Feb 28;11(3):150. doi: 10.3390/insects11030150. Insects. 2020. PMID: 32121261 Free PMC article.
-
Insect Cuticular Chitin Contributes to Form and Function.Curr Pharm Des. 2020;26(29):3530-3545. doi: 10.2174/1381612826666200523175409. Curr Pharm Des. 2020. PMID: 32445445 Free PMC article.
-
Molecular Characterization of Chitin Synthase Gene in Tetranychus cinnabarinus (Boisduval) and Its Response to Sublethal Concentrations of an Insecticide.Insects. 2021 May 28;12(6):501. doi: 10.3390/insects12060501. Insects. 2021. PMID: 34071207 Free PMC article.
-
Identification and Functional Study of Chitin Metabolism and Detoxification-Related Genes in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae) Based on Transcriptome Analysis.Int J Mol Sci. 2020 Mar 10;21(5):1904. doi: 10.3390/ijms21051904. Int J Mol Sci. 2020. PMID: 32164390 Free PMC article.
References
-
- Chapman RF. The Insects: Structure and Function. Cambridge, MA: Harvard Univ Press; 1998.
-
- Kramer KJ, Muthukrishnan S. Chitin metabolism in insects. In: Gilbert L.I, latrou K, Gill S, editors. Comprehensive Molecular Insect Science; Vol. 4, Biochemistry and Molecular Biology, Chapter 3. Oxford, UK: Elsevier Press; 2005. pp. 111–144.
-
- Merzendorfer H. Insect chitin synthases: a review. J. Comp. Physiol. 2006;176:1–15. - PubMed
-
- Merzendorfer H, Zimoch L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and Chitinases. J. Exp. Bio. 2003;206:4393–4412. - PubMed
-
- Arakane Y, Specht CA, Kramer KJ, Muthukrishnan S, Beeman RW. Chitin synthases are required for survival, fecundity and egg hatch in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2008;38:959–962. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

