Structural basis of fluorescence quenching in caspase activatable-GFP
- PMID: 23139158
- PMCID: PMC3595455
- DOI: 10.1002/pro.2188
Structural basis of fluorescence quenching in caspase activatable-GFP
Abstract
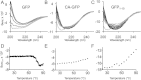
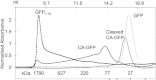
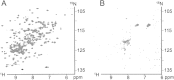
Apoptosis is critical for organismal homeostasis and a wide variety of diseases. Caspases are the ultimate executors of the apoptotic programmed cell death pathway. As caspases play such a central role in apoptosis, there is significant demand for technologies to monitor caspase function. We recently developed a caspase activatable-GFP (CA-GFP) reporter. CA-GFP is unique due to its "dark" state, where chromophore maturation of the GFP is inhibited by the presence of a C-terminal peptide. Here we show that chromophore maturation is prevented because CA-GFP does not fold into the robust β-barrel of GFP until the peptide has been cleaved by active caspase. Both CA-GFP and GFP₁₋₁₀ , a split form of GFP lacking the 11th strand, have similar secondary structure, different from mature GFP. A similar susceptibility to proteolytic digestion indicates that this shared structure is not the robust, fully formed GFP β-barrel. We have developed a model that suggests that as CA-GFP is translated in vivo it follows the same folding path as wild-type GFP; however, the presence of the appended peptide does not allow CA-GFP to form the barrel of the fully matured GFP. CA-GFP is therefore held in a "pro-folding" intermediate state until the peptide is released, allowing it to continue folding into the mature barrel geometry. This new understanding of the structural basis of the dark state of the CA-GFP reporter will enable manipulation of this mechanism in the development of reporter systems for any number of cellular processes involving proteases and potentially other enzymes.
Copyright © 2013 The Protein Society.
Figures



Similar articles
-
A tunable, modular approach to fluorescent protease-activated reporters.Biophys J. 2013 Apr 2;104(7):1605-14. doi: 10.1016/j.bpj.2013.01.058. Biophys J. 2013. PMID: 23561537 Free PMC article.
-
Mechanism of a genetically encoded dark-to-bright reporter for caspase activity.J Biol Chem. 2011 Jul 15;286(28):24977-86. doi: 10.1074/jbc.M111.221648. Epub 2011 May 10. J Biol Chem. 2011. PMID: 21558267 Free PMC article.
-
The rough energy landscape of superfolder GFP is linked to the chromophore.J Mol Biol. 2007 Oct 19;373(2):476-90. doi: 10.1016/j.jmb.2007.07.071. Epub 2007 Aug 15. J Mol Biol. 2007. PMID: 17822714 Free PMC article.
-
Ac-rkkrrorrrGK(QSY21)DEVDAPC(Alexa Fluor 647)-NH2.2008 Jan 23 [updated 2008 Apr 22]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. 2008 Jan 23 [updated 2008 Apr 22]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. PMID: 20641692 Free Books & Documents. Review.
-
Measuring caspase activity in vivo.Methods Enzymol. 2014;544:251-69. doi: 10.1016/B978-0-12-417158-9.00010-8. Methods Enzymol. 2014. PMID: 24974293 Review.
Cited by
-
Development of a fluorescence-based cellular apoptosis reporter.Methods Appl Fluoresc. 2018 Oct 24;7(1):015001. doi: 10.1088/2050-6120/aae6f8. Methods Appl Fluoresc. 2018. PMID: 30353887 Free PMC article.
-
A tunable, modular approach to fluorescent protease-activated reporters.Biophys J. 2013 Apr 2;104(7):1605-14. doi: 10.1016/j.bpj.2013.01.058. Biophys J. 2013. PMID: 23561537 Free PMC article.
-
A fluorescence-activatable reporter of flavivirus NS2B-NS3 protease activity enables live imaging of infection in single cells and viral plaques.J Biol Chem. 2020 Feb 21;295(8):2212-2226. doi: 10.1074/jbc.RA119.011319. Epub 2020 Jan 9. J Biol Chem. 2020. PMID: 31919100 Free PMC article.
-
The Use of ex Vivo Rodent Platforms in Neuroscience Translational Research With Attention to the 3Rs Philosophy.Front Vet Sci. 2018 Jul 19;5:164. doi: 10.3389/fvets.2018.00164. eCollection 2018. Front Vet Sci. 2018. PMID: 30073174 Free PMC article. Review.
-
Live Cell Reporter Systems for Positive-Sense Single Strand RNA Viruses.Appl Biochem Biotechnol. 2016 Apr;178(8):1567-85. doi: 10.1007/s12010-015-1968-5. Epub 2016 Jan 4. Appl Biochem Biotechnol. 2016. PMID: 26728654 Free PMC article. Review.
References
-
- I G, AD H, Regan L. Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J Am Chem Soc. 2000;122:5658–5659.
-
- Hu C, Chinenov Y, Kerppola T. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. - PubMed
-
- Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nature Biotech. 2004;23:102–107. - PubMed
-
- Wood TI, Barondeau Dp, Hitomi C, Kassmann CJ, Trainer JA, Getzoff ED. Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis. Biochemistry. 2005;44:16211–16220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

