Genetic incorporation of twelve meta-substituted phenylalanine derivatives using a single pyrrolysyl-tRNA synthetase mutant
- PMID: 23138887
- PMCID: PMC3574229
- DOI: 10.1021/cb300512r
Genetic incorporation of twelve meta-substituted phenylalanine derivatives using a single pyrrolysyl-tRNA synthetase mutant
Abstract
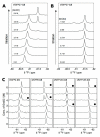
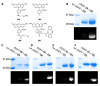
When coexpressed with its cognate amber suppressing tRNACUAPyl(CUA), a pyrrolysyltRNA synthetase mutant N346A/C348A is able to genetically incorporate 12 meta-substituted phenylalanine derivatives into proteins site-specifically at amber mutation sites in Escherichia coli. These genetically encoded noncanonical amino acids resemble phenylalanine in size and contain diverse bioorthogonal functional groups such as halide, trifluoromethyl, nitrile, nitro,ketone, alkyne, and azide moieties. The genetic installation of these functional groups in proteins provides multiple ways to site-selectively label proteins with biophysical and biochemical probes for their functional investigations. We demonstrate that a genetically incorporated trifluoromethyl group can be used as a sensitive 19F NMR probe to study protein folding/unfolding, and that genetically incorporated reactive functional groups such as ketone,alkyne, and azide moieties can be applied to site-specifically label proteins with fluorescent probes. This critical discovery allows the synthesis of proteins with diverse bioorthogonal functional groups for a variety of basic studies and biotechnology development using a single recombinant expression system.
Figures



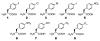
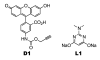
Similar articles
-
The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives.Mol Biosyst. 2011 Mar;7(3):714-7. doi: 10.1039/c0mb00217h. Epub 2011 Jan 14. Mol Biosyst. 2011. PMID: 21234492
-
A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum.J Am Chem Soc. 2012 Feb 15;134(6):2950-3. doi: 10.1021/ja211972x. Epub 2012 Feb 6. J Am Chem Soc. 2012. PMID: 22289053 Free PMC article.
-
Genetic incorporation of histidine derivatives using an engineered pyrrolysyl-tRNA synthetase.ACS Chem Biol. 2014 May 16;9(5):1092-6. doi: 10.1021/cb500032c. Epub 2014 Mar 17. ACS Chem Biol. 2014. PMID: 24506189 Free PMC article.
-
tRNAPyl: Structure, function, and applications.RNA Biol. 2018;15(4-5):441-452. doi: 10.1080/15476286.2017.1356561. Epub 2017 Sep 13. RNA Biol. 2018. PMID: 28837402 Free PMC article. Review.
-
An expanding genetic code.Trends Genet. 2004 Dec;20(12):625-30. doi: 10.1016/j.tig.2004.09.013. Trends Genet. 2004. PMID: 15522458 Review.
Cited by
-
Efficient Reassignment of a Frequent Serine Codon in Wild-Type Escherichia coli.ACS Synth Biol. 2016 Feb 19;5(2):163-71. doi: 10.1021/acssynbio.5b00197. Epub 2015 Nov 20. ACS Synth Biol. 2016. PMID: 26544153 Free PMC article.
-
Update of the Pyrrolysyl-tRNA Synthetase/tRNAPyl Pair and Derivatives for Genetic Code Expansion.J Bacteriol. 2023 Feb 22;205(2):e0038522. doi: 10.1128/jb.00385-22. Epub 2023 Jan 25. J Bacteriol. 2023. PMID: 36695595 Free PMC article. Review.
-
A Designed, Highly Efficient Pyrrolysyl-tRNA Synthetase Mutant Binds o-Chlorophenylalanine Using Two Halogen Bonds.J Mol Biol. 2022 Apr 30;434(8):167534. doi: 10.1016/j.jmb.2022.167534. Epub 2022 Mar 9. J Mol Biol. 2022. PMID: 35278475 Free PMC article.
-
Chemically-defined lactose-based autoinduction medium for site-specific incorporation of non-canonical amino acids into proteins.RSC Adv. 2018 Jul 17;8(45):25558-25567. doi: 10.1039/c8ra04359k. RSC Adv. 2018. PMID: 30713681 Free PMC article.
-
Visualizing an Allosteric Intermediate Using CuAAC Stabilization of an NMR Mixed Labeled Dimer.ACS Chem Biol. 2021 Dec 17;16(12):2766-2775. doi: 10.1021/acschembio.1c00617. Epub 2021 Nov 16. ACS Chem Biol. 2021. PMID: 34784173 Free PMC article.
References
-
- Roberts MJ, Bentley MD, Harris JM. Chemistry for peptide and protein PEGylation. Adv Drug Deliv Rev. 2002;54:459–476. - PubMed
-
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. - PubMed
-
- Zhang M, Lin S, Song X, Liu J, Fu Y, Ge X, Fu X, Chang Z, Chen PR. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat Chem Biol. 2011;7:671–677. - PubMed
-
- Chin JW, Schultz PG. In vivo photocrosslinking with unnatural amino Acid mutagenesis. Chembiochem. 2002;3:1135–1137. - PubMed
-
- Jones DH, Cellitti SE, Hao X, Zhang Q, Jahnz M, Summerer D, Schultz PG, Uno T, Geierstanger BH. Site-specific labeling of proteins with NMR-active unnatural amino acids. J Biomol NMR. 2010;46:89–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

