Long non-coding RNAs function annotation: a global prediction method based on bi-colored networks
- PMID: 23132350
- PMCID: PMC3554231
- DOI: 10.1093/nar/gks967
Long non-coding RNAs function annotation: a global prediction method based on bi-colored networks
Abstract
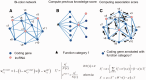
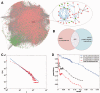
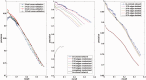
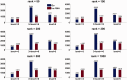
More and more evidences demonstrate that the long non-coding RNAs (lncRNAs) play many key roles in diverse biological processes. There is a critical need to annotate the functions of increasing available lncRNAs. In this article, we try to apply a global network-based strategy to tackle this issue for the first time. We develop a bi-colored network based global function predictor, long non-coding RNA global function predictor ('lnc-GFP'), to predict probable functions for lncRNAs at large scale by integrating gene expression data and protein interaction data. The performance of lnc-GFP is evaluated on protein-coding and lncRNA genes. Cross-validation tests on protein-coding genes with known function annotations indicate that our method can achieve a precision up to 95%, with a suitable parameter setting. Among the 1713 lncRNAs in the bi-colored network, the 1625 (94.9%) lncRNAs in the maximum connected component are all functionally characterized. For the lncRNAs expressed in mouse embryo stem cells and neuronal cells, the inferred putative functions by our method highly match those in the known literature.
Figures
Similar articles
-
Predicting the functions of long noncoding RNAs using RNA-seq based on Bayesian network.Biomed Res Int. 2015;2015:839590. doi: 10.1155/2015/839590. Epub 2015 Feb 28. Biomed Res Int. 2015. PMID: 25815337 Free PMC article.
-
KATZLGO: Large-Scale Prediction of LncRNA Functions by Using the KATZ Measure Based on Multiple Networks.IEEE/ACM Trans Comput Biol Bioinform. 2019 Mar-Apr;16(2):407-416. doi: 10.1109/TCBB.2017.2704587. Epub 2017 May 16. IEEE/ACM Trans Comput Biol Bioinform. 2019. PMID: 28534780
-
Gene Ontology-based function prediction of long non-coding RNAs using bi-random walk.BMC Med Genomics. 2018 Nov 20;11(Suppl 5):99. doi: 10.1186/s12920-018-0414-2. BMC Med Genomics. 2018. PMID: 30453964 Free PMC article.
-
Computational approaches towards understanding human long non-coding RNA biology.Bioinformatics. 2015 Jul 15;31(14):2241-51. doi: 10.1093/bioinformatics/btv148. Epub 2015 Mar 15. Bioinformatics. 2015. PMID: 25777523 Review.
-
Long non-coding RNAs in human early embryonic development and their potential in ART.Hum Reprod Update. 2016 Dec;23(1):19-40. doi: 10.1093/humupd/dmw035. Epub 2016 Sep 21. Hum Reprod Update. 2016. PMID: 27655590 Review.
Cited by
-
Predicting the functions of long noncoding RNAs using RNA-seq based on Bayesian network.Biomed Res Int. 2015;2015:839590. doi: 10.1155/2015/839590. Epub 2015 Feb 28. Biomed Res Int. 2015. PMID: 25815337 Free PMC article.
-
A novel heterogeneous network-based method for drug response prediction in cancer cell lines.Sci Rep. 2018 Feb 20;8(1):3355. doi: 10.1038/s41598-018-21622-4. Sci Rep. 2018. PMID: 29463808 Free PMC article.
-
Towards a complete map of the human long non-coding RNA transcriptome.Nat Rev Genet. 2018 Sep;19(9):535-548. doi: 10.1038/s41576-018-0017-y. Nat Rev Genet. 2018. PMID: 29795125 Free PMC article. Review.
-
Comprehensive analysis of lncRNA-mRNA co-expression patterns identifies immune-associated lncRNA biomarkers in ovarian cancer malignant progression.Sci Rep. 2015 Dec 3;5:17683. doi: 10.1038/srep17683. Sci Rep. 2015. PMID: 26631459 Free PMC article.
-
Annotation and functional characterization of long noncoding RNAs deregulated in pancreatic adenocarcinoma.Cell Oncol (Dordr). 2022 Jun;45(3):479-504. doi: 10.1007/s13402-022-00678-5. Epub 2022 May 14. Cell Oncol (Dordr). 2022. PMID: 35567709
References
-
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. - PubMed
-
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2003;36:40–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

