Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments
- PMID: 23129641
- PMCID: PMC3523859
- DOI: 10.1073/pnas.1208618109
Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments
Abstract
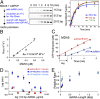
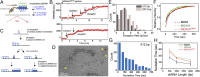
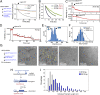
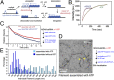
The viral sensor MDA5 distinguishes between cellular and viral dsRNAs by length-dependent recognition in the range of ~0.5-7 kb. The ability to discriminate dsRNA length at this scale sets MDA5 apart from other dsRNA receptors of the immune system. We have shown previously that MDA5 forms filaments along dsRNA that disassemble upon ATP hydrolysis. Here, we demonstrate that filament formation alone is insufficient to explain its length specificity, because the intrinsic affinity of MDA5 for dsRNA depends only moderately on dsRNA length. Instead, MDA5 uses a combination of end disassembly and slow nucleation kinetics to "discard" short dsRNA rapidly and to suppress rebinding. In contrast, filaments on long dsRNA cycle between partial end disassembly and elongation, bypassing nucleation steps. MDA5 further uses this repetitive cycle of assembly and disassembly processes to repair filament discontinuities, which often are present because of multiple, internal nucleation events, and to generate longer, continuous filaments that more accurately reflect the length of the underlying dsRNA scaffold. Because the length of the continuous filament determines the stability of the MDA5-dsRNA interaction, the mechanism proposed here provides an explanation for how MDA5 uses filament assembly and disassembly dynamics to discriminate between self vs. nonself dsRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition.Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21010-5. doi: 10.1073/pnas.1113651108. Epub 2011 Dec 12. Proc Natl Acad Sci U S A. 2011. PMID: 22160685 Free PMC article.
-
Contrasting functions of ATP hydrolysis by MDA5 and LGP2 in viral RNA sensing.J Biol Chem. 2024 Mar;300(3):105711. doi: 10.1016/j.jbc.2024.105711. Epub 2024 Feb 1. J Biol Chem. 2024. PMID: 38309507 Free PMC article.
-
MDA5-filament, dynamics and disease.Curr Opin Virol. 2015 Jun;12:20-5. doi: 10.1016/j.coviro.2015.01.011. Epub 2015 Feb 9. Curr Opin Virol. 2015. PMID: 25676875 Free PMC article. Review.
-
The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly.Mol Cell. 2014 Sep 4;55(5):771-81. doi: 10.1016/j.molcel.2014.07.003. Epub 2014 Aug 7. Mol Cell. 2014. PMID: 25127512 Free PMC article.
-
To "Z" or not to "Z": Z-RNA, self-recognition, and the MDA5 helicase.PLoS Genet. 2021 May 13;17(5):e1009513. doi: 10.1371/journal.pgen.1009513. eCollection 2021 May. PLoS Genet. 2021. PMID: 33983939 Free PMC article. Review.
Cited by
-
High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5.Nucleic Acids Res. 2015 Jan;43(2):1216-30. doi: 10.1093/nar/gku1329. Epub 2014 Dec 24. Nucleic Acids Res. 2015. PMID: 25539915 Free PMC article.
-
Unraveling blunt-end RNA binding and ATPase-driven translocation activities of the RIG-I family helicase LGP2.Nucleic Acids Res. 2024 Jan 11;52(1):355-369. doi: 10.1093/nar/gkad1106. Nucleic Acids Res. 2024. PMID: 38015453 Free PMC article.
-
An engineered T7 RNA polymerase that produces mRNA free of immunostimulatory byproducts.Nat Biotechnol. 2023 Apr;41(4):560-568. doi: 10.1038/s41587-022-01525-6. Epub 2022 Nov 10. Nat Biotechnol. 2023. PMID: 36357718 Free PMC article.
-
Filament-like Assemblies of Intracellular Nucleic Acid Sensors: Commonalities and Differences.Mol Cell. 2019 Oct 17;76(2):243-254. doi: 10.1016/j.molcel.2019.09.023. Mol Cell. 2019. PMID: 31626748 Free PMC article. Review.
-
Cellular origins of dsRNA, their recognition and consequences.Nat Rev Mol Cell Biol. 2022 Apr;23(4):286-301. doi: 10.1038/s41580-021-00430-1. Epub 2021 Nov 23. Nat Rev Mol Cell Biol. 2022. PMID: 34815573 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

